bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2100_orf1
Length=96
Score E
Sequences producing significant alignments: (Bits) Value
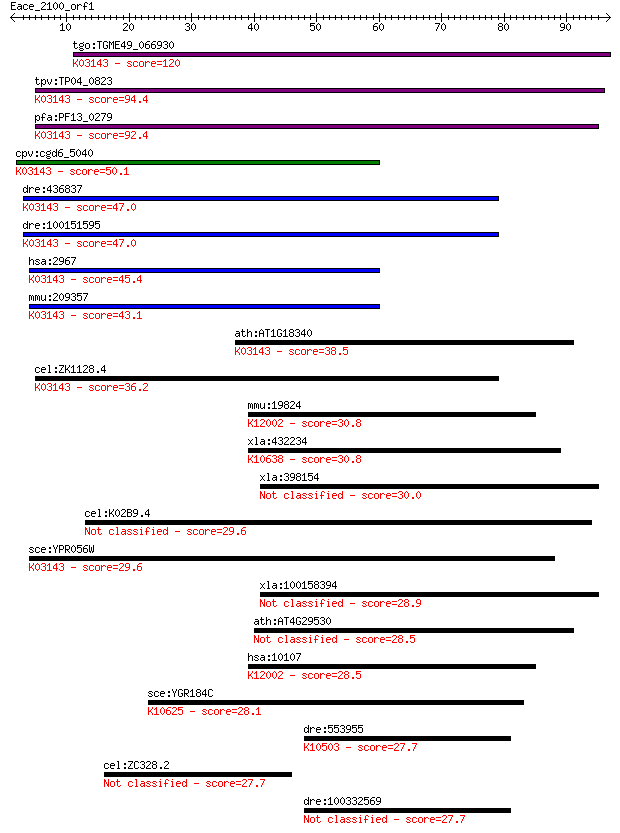
tgo:TGME49_066930 hypothetical protein ; K03143 transcription ... 120 8e-28
tpv:TP04_0823 hypothetical protein; K03143 transcription initi... 94.4 8e-20
pfa:PF13_0279 conserved Plasmodium protein, unknown function; ... 92.4 3e-19
cpv:cgd6_5040 transcription factor ; K03143 transcription init... 50.1 2e-06
dre:436837 gtf2h3, fc26a07, wu:fc26a07, zgc:92759; general tra... 47.0 1e-05
dre:100151595 gtf2h3-2; general transcription factor IIH, poly... 47.0 2e-05
hsa:2967 GTF2H3, BTF2, TFB4, TFIIH; general transcription fact... 45.4 4e-05
mmu:209357 Gtf2h3, 34kDa, 5033417D07Rik, BTF2, BTF2_p34, C7300... 43.1 2e-04
ath:AT1G18340 basal transcription factor complex subunit-relat... 38.5 0.006
cel:ZK1128.4 hypothetical protein; K03143 transcription initia... 36.2 0.028
mmu:19824 Trim10, AI324236, BB139825, Herf1, Rnf9; tripartite ... 30.8 1.0
xla:432234 uhrf1; ubiquitin-like with PHD and ring finger doma... 30.8 1.2
xla:398154 gatad2a; GATA zinc finger domain containing 2A 30.0 2.1
cel:K02B9.4 elt-3; Erythroid-Like Transcription factor family ... 29.6 2.4
sce:YPR056W TFB4; Subunit of TFIIH complex, involved in transc... 29.6 2.7
xla:100158394 hypothetical protein LOC100158394 28.9 4.0
ath:AT4G29530 2,3-diketo-5-methylthio-1-phosphopentane phospha... 28.5 5.1
hsa:10107 TRIM10, HERF1, MGC141979, RFB30, RNF9; tripartite mo... 28.5 5.4
sce:YGR184C UBR1, PTR1; E3 ubiquitin ligase (N-recognin), form... 28.1 7.9
dre:553955 zbtb24, zfp450, zgc:194556; zinc finger and BTB dom... 27.7 8.2
cel:ZC328.2 hypothetical protein 27.7 8.7
dre:100332569 zinc finger protein 333-like 27.7 8.9
> tgo:TGME49_066930 hypothetical protein ; K03143 transcription
initiation factor TFIIH subunit 3
Length=547
Score = 120 bits (302), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 56/88 (63%), Positives = 68/88 (77%), Gaps = 2/88 (2%)
Query 11 FWFLPPPSLRPSVAAVSIHH-KTNAGVCFCHHNPVDLCFICSCCLAIYCSET-SDNGKER 68
FW P SLRP++AA+S+H ++N VCFCHH PV++C ICSCCLAIYCSE + GKER
Sbjct 460 FWIFPSMSLRPAIAALSVHRGRSNTAVCFCHHKPVEVCCICSCCLAIYCSEKDAQTGKER 519
Query 69 ISCDVCKSRFAKGLLKAKLAASVDASKY 96
ISCDVCKSRF++GLLK K+A VD Y
Sbjct 520 ISCDVCKSRFSRGLLKNKMAGGVDLPNY 547
> tpv:TP04_0823 hypothetical protein; K03143 transcription initiation
factor TFIIH subunit 3
Length=348
Score = 94.4 bits (233), Expect = 8e-20, Method: Composition-based stats.
Identities = 40/91 (43%), Positives = 54/91 (59%), Gaps = 0/91 (0%)
Query 5 LLQLLVFWFLPPPSLRPSVAAVSIHHKTNAGVCFCHHNPVDLCFICSCCLAIYCSETSDN 64
L QL+ FW+LP + ++ N G+C+CH+ VD+ ++C CC A+YCSE D
Sbjct 257 LSQLITFWYLPSEGMSELLSTNLSFDFGNVGICYCHYKSVDVSYLCPCCFAVYCSEIDDK 316
Query 65 GKERISCDVCKSRFAKGLLKAKLAASVDASK 95
GK RI C VC SR + L+K KLA D SK
Sbjct 317 GKYRIICMVCGSRLTRKLIKQKLATEADFSK 347
> pfa:PF13_0279 conserved Plasmodium protein, unknown function;
K03143 transcription initiation factor TFIIH subunit 3
Length=326
Score = 92.4 bits (228), Expect = 3e-19, Method: Composition-based stats.
Identities = 41/91 (45%), Positives = 59/91 (64%), Gaps = 1/91 (1%)
Query 5 LLQLLVFWFLPPPSLRPSVAAVSIHHKTNAGVCFCHHNPVDLCFICSCCLAIYCSETS-D 63
L Q ++FWFLP + R + ++ TN VC CH+ +D+ +ICSCCLAIYCSE +
Sbjct 234 LTQTIIFWFLPSVNTRKYFSNTYLNEDTNIAVCTCHNKQIDIAYICSCCLAIYCSEKNLQ 293
Query 64 NGKERISCDVCKSRFAKGLLKAKLAASVDAS 94
K+R+SC +CK+RF K LL+ K + +D S
Sbjct 294 TNKDRLSCAICKTRFTKSLLRNKHVSDLDFS 324
> cpv:cgd6_5040 transcription factor ; K03143 transcription initiation
factor TFIIH subunit 3
Length=286
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 34/59 (57%), Gaps = 1/59 (1%)
Query 2 DIALLQLLVFWFLPPPSLRPSV-AAVSIHHKTNAGVCFCHHNPVDLCFICSCCLAIYCS 59
D L L+F LP R + +++ +T VCFCHH +++ ++CS CL+I+CS
Sbjct 217 DQGLTPFLIFHLLPSIQAREEIFISINKTKQTGLAVCFCHHQKIEIGYVCSSCLSIFCS 275
> dre:436837 gtf2h3, fc26a07, wu:fc26a07, zgc:92759; general transcription
factor IIH, polypeptide 3; K03143 transcription
initiation factor TFIIH subunit 3
Length=296
Score = 47.0 bits (110), Expect = 1e-05, Method: Composition-based stats.
Identities = 23/76 (30%), Positives = 38/76 (50%), Gaps = 7/76 (9%)
Query 3 IALLQLLVFWFLPPPSLRPSVAAVSIHHKTNAGVCFCHHNPVDLCFICSCCLAIYCSETS 62
+AL Q L++ FLP R + H CFCH N +++ ++CS CL+I+C+ +
Sbjct 209 VALTQYLLWAFLPDAEQRSQLLLPPPVHVDYRAACFCHRNLIEIGYVCSVCLSIFCNFSP 268
Query 63 DNGKERISCDVCKSRF 78
C C++ F
Sbjct 269 -------ICTTCETAF 277
> dre:100151595 gtf2h3-2; general transcription factor IIH, polypeptide
3; K03143 transcription initiation factor TFIIH subunit
3
Length=296
Score = 47.0 bits (110), Expect = 2e-05, Method: Composition-based stats.
Identities = 23/76 (30%), Positives = 38/76 (50%), Gaps = 7/76 (9%)
Query 3 IALLQLLVFWFLPPPSLRPSVAAVSIHHKTNAGVCFCHHNPVDLCFICSCCLAIYCSETS 62
+AL Q L++ FLP R + H CFCH N +++ ++CS CL+I+C+ +
Sbjct 209 VALTQYLLWVFLPDAEQRSQLLLPPPVHVDYRAACFCHRNLIEIGYVCSVCLSIFCNFSP 268
Query 63 DNGKERISCDVCKSRF 78
C C++ F
Sbjct 269 -------ICTTCETAF 277
> hsa:2967 GTF2H3, BTF2, TFB4, TFIIH; general transcription factor
IIH, polypeptide 3, 34kDa; K03143 transcription initiation
factor TFIIH subunit 3
Length=308
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 32/56 (57%), Gaps = 0/56 (0%)
Query 4 ALLQLLVFWFLPPPSLRPSVAAVSIHHKTNAGVCFCHHNPVDLCFICSCCLAIYCS 59
+LLQ L++ FLP R + H CFCH N +++ ++CS CL+I+C+
Sbjct 222 SLLQYLLWVFLPDQDQRSQLILPPPVHVDYRAACFCHRNLIEIGYVCSVCLSIFCN 277
> mmu:209357 Gtf2h3, 34kDa, 5033417D07Rik, BTF2, BTF2_p34, C730029A10,
D5Ertd679e, TFIIH; general transcription factor IIH,
polypeptide 3; K03143 transcription initiation factor TFIIH
subunit 3
Length=309
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 32/56 (57%), Gaps = 0/56 (0%)
Query 4 ALLQLLVFWFLPPPSLRPSVAAVSIHHKTNAGVCFCHHNPVDLCFICSCCLAIYCS 59
+LLQ L++ FLP R + H CFCH + +++ ++CS CL+I+C+
Sbjct 223 SLLQYLLWVFLPDQDQRSQLILPPPIHVDYRAACFCHRSLIEIGYVCSVCLSIFCN 278
> ath:AT1G18340 basal transcription factor complex subunit-related;
K03143 transcription initiation factor TFIIH subunit 3
Length=301
Score = 38.5 bits (88), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 19/54 (35%), Positives = 27/54 (50%), Gaps = 7/54 (12%)
Query 37 CFCHHNPVDLCFICSCCLAIYCSETSDNGKERISCDVCKSRFAKGLLKAKLAAS 90
CFCH +D+ +ICS CL+I+C + C C S F + L +AS
Sbjct 246 CFCHKKTIDMGYICSVCLSIFC-------EHHKKCSTCGSVFGQSKLDDASSAS 292
> cel:ZK1128.4 hypothetical protein; K03143 transcription initiation
factor TFIIH subunit 3
Length=345
Score = 36.2 bits (82), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 35/74 (47%), Gaps = 7/74 (9%)
Query 5 LLQLLVFWFLPPPSLRPSVAAVSIHHKTNAGVCFCHHNPVDLCFICSCCLAIYCSETSDN 64
LL++L+ L P+ R + +S + C CHH V ++CS CL++ C T
Sbjct 262 LLKILMTNMLTDPTHRAVFSKLSHNSVDYRASCACHHQLVSSGWVCSICLSVLCQYTP-- 319
Query 65 GKERISCDVCKSRF 78
C VCK+ F
Sbjct 320 -----ICKVCKAAF 328
> mmu:19824 Trim10, AI324236, BB139825, Herf1, Rnf9; tripartite
motif-containing 10; K12002 tripartite motif-containing protein
10
Length=489
Score = 30.8 bits (68), Expect = 1.0, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 19/46 (41%), Gaps = 6/46 (13%)
Query 39 CHHNPVDLCFICSCCLAIYCSETSDNGKERISCDVCKSRFAKGLLK 84
C HN C CL YC +E +SC +CK F G +
Sbjct 31 CGHN------FCRGCLTRYCEIPGPESEESLSCPLCKEPFRPGSFR 70
> xla:432234 uhrf1; ubiquitin-like with PHD and ring finger domains
1 (EC:6.3.2.19); K10638 E3 ubiquitin-protein ligase UHRF1
[EC:6.3.2.19]
Length=772
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 23/50 (46%), Gaps = 4/50 (8%)
Query 39 CHHNPVDLCFICSCCLAIYCSETSDNGKERISCDVCKSRFAKGLLKAKLA 88
C NP C +C+CC+ C D K+ + CD C F LK L+
Sbjct 303 CKDNPKRACRMCACCI---CGGKQDPEKQLL-CDECDLAFHIYCLKPPLS 348
> xla:398154 gatad2a; GATA zinc finger domain containing 2A
Length=420
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 17/54 (31%), Positives = 24/54 (44%), Gaps = 4/54 (7%)
Query 41 HNPVDLCFICSCCLAIYCSETSDNGKERISCDVCKSRFAKGLLKAKLAASVDAS 94
H P +IC+ C + S I CDVC S K +LKA+ + A+
Sbjct 231 HEP----YICAKCKTDFTSRWRQEKNGTIMCDVCMSSNQKKVLKAEHTTRLKAA 280
> cel:K02B9.4 elt-3; Erythroid-Like Transcription factor family
member (elt-3)
Length=226
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 31/81 (38%), Gaps = 7/81 (8%)
Query 13 FLPPPSLRPSVAAVSIHHKTNAGVCFCHHNPVDLCFICSCCLAIYCSETSDNGKERISCD 72
F PPP P VA K A V CH N ICS C + NG+ + C+
Sbjct 123 FTPPPQ-DPLVAEPKPMKKRMAAVQ-CHQN-----SICSNCKTRETTLWRRNGEGGVECN 175
Query 73 VCKSRFAKGLLKAKLAASVDA 93
C F K K L+ D
Sbjct 176 ACNLYFRKNNRKRPLSLRKDG 196
> sce:YPR056W TFB4; Subunit of TFIIH complex, involved in transcription
initiation, similar to 34 kDa subunit of human TFIIH;
interacts with Ssl1p; K03143 transcription initiation factor
TFIIH subunit 3
Length=338
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 26/87 (29%), Positives = 38/87 (43%), Gaps = 12/87 (13%)
Query 4 ALLQLLVFWFLPPPSLRPSVAAV---SIHHKTNAGVCFCHHNPVDLCFICSCCLAIYCSE 60
L+Q L PSLRP + S+ +T+ C+ V + FICS CL +
Sbjct 243 GLIQYLATAMFIDPSLRPIIVKPNHGSVDFRTS---CYLTGRVVAVGFICSVCLCVLSII 299
Query 61 TSDNGKERISCDVCKSRFAKGLLKAKL 87
N C C S+F + ++ AKL
Sbjct 300 PPGN-----KCPACDSQFDEHVI-AKL 320
> xla:100158394 hypothetical protein LOC100158394
Length=555
Score = 28.9 bits (63), Expect = 4.0, Method: Composition-based stats.
Identities = 17/54 (31%), Positives = 23/54 (42%), Gaps = 4/54 (7%)
Query 41 HNPVDLCFICSCCLAIYCSETSDNGKERISCDVCKSRFAKGLLKAKLAASVDAS 94
H P +IC+ C + S I CDVC S K LKA+ + A+
Sbjct 368 HEP----YICAKCKTDFTSRWRQEKNGTIMCDVCMSSNQKKALKAEHTTRLKAA 417
> ath:AT4G29530 2,3-diketo-5-methylthio-1-phosphopentane phosphatase
family
Length=245
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 29/64 (45%), Gaps = 19/64 (29%)
Query 40 HHNPVDLCFICSCCLAIYCSETS--DNGKERI-----------SCDVCKSRFAKGLLKAK 86
HH+ VD C IY + TS DNG RI SC++C S KGL+
Sbjct 107 HHDLVD------CFSEIYTNPTSLDDNGNLRILPYHSDALPPHSCNLCPSNLCKGLVMDH 160
Query 87 LAAS 90
L AS
Sbjct 161 LRAS 164
> hsa:10107 TRIM10, HERF1, MGC141979, RFB30, RNF9; tripartite
motif containing 10; K12002 tripartite motif-containing protein
10
Length=481
Score = 28.5 bits (62), Expect = 5.4, Method: Composition-based stats.
Identities = 14/46 (30%), Positives = 19/46 (41%), Gaps = 6/46 (13%)
Query 39 CHHNPVDLCFICSCCLAIYCSETSDNGKERISCDVCKSRFAKGLLK 84
C HN C CL YC + +E +C +CK F G +
Sbjct 31 CGHN------FCRACLTRYCEIPGPDLEESPTCPLCKEPFRPGSFR 70
> sce:YGR184C UBR1, PTR1; E3 ubiquitin ligase (N-recognin), forms
heterodimer with Rad6p to ubiquitinate substrates in the
N-end rule pathway; regulates peptide transport via Cup9p ubiquitination;
mutation in human UBR1 causes Johansson-Blizzard
Syndrome (JBS) (EC:6.3.2.-); K10625 E3 ubiquitin-protein
ligase UBR1 [EC:6.3.2.19]
Length=1950
Score = 28.1 bits (61), Expect = 7.9, Method: Composition-based stats.
Identities = 17/64 (26%), Positives = 26/64 (40%), Gaps = 5/64 (7%)
Query 23 VAAVSIHHKTNAGVCFCHHNPVDLCFIC----SCCLAIYCSETSDNGKERISCDVCKSRF 78
+ V H N G F P+ C C +C L I+C D+ + D+C + F
Sbjct 112 IGDVHKHTGRNCGRKFKIGEPLYRCHECGCDDTCVLCIHCFNPKDHVNHHVCTDIC-TEF 170
Query 79 AKGL 82
G+
Sbjct 171 TSGI 174
> dre:553955 zbtb24, zfp450, zgc:194556; zinc finger and BTB domain
containing 24; K10503 zinc finger and BTB domain-containing
protein 24
Length=672
Score = 27.7 bits (60), Expect = 8.2, Method: Composition-based stats.
Identities = 12/40 (30%), Positives = 18/40 (45%), Gaps = 7/40 (17%)
Query 48 FICSCCLAIYCS-------ETSDNGKERISCDVCKSRFAK 80
++C C + E S GK+ SC +CK FA+
Sbjct 405 YVCKVCDKTFSDPSARRRHEVSHTGKKTFSCSICKVSFAR 444
> cel:ZC328.2 hypothetical protein
Length=386
Score = 27.7 bits (60), Expect = 8.7, Method: Composition-based stats.
Identities = 12/31 (38%), Positives = 18/31 (58%), Gaps = 1/31 (3%)
Query 16 PPSLRPSVAAVSIHHKTNA-GVCFCHHNPVD 45
PP L P AA++I H+ +A + CHH +
Sbjct 167 PPGLSPGGAALNIEHEKSAFEIVSCHHQSAE 197
> dre:100332569 zinc finger protein 333-like
Length=678
Score = 27.7 bits (60), Expect = 8.9, Method: Composition-based stats.
Identities = 12/40 (30%), Positives = 18/40 (45%), Gaps = 7/40 (17%)
Query 48 FICSCCLAIYCS-------ETSDNGKERISCDVCKSRFAK 80
++C C + E S GK+ SC +CK FA+
Sbjct 408 YVCKVCDKTFSDPSARRRHEVSHTGKKTFSCSICKVSFAR 447
Lambda K H
0.327 0.138 0.462
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2060454524
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40