bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2095_orf2
Length=104
Score E
Sequences producing significant alignments: (Bits) Value
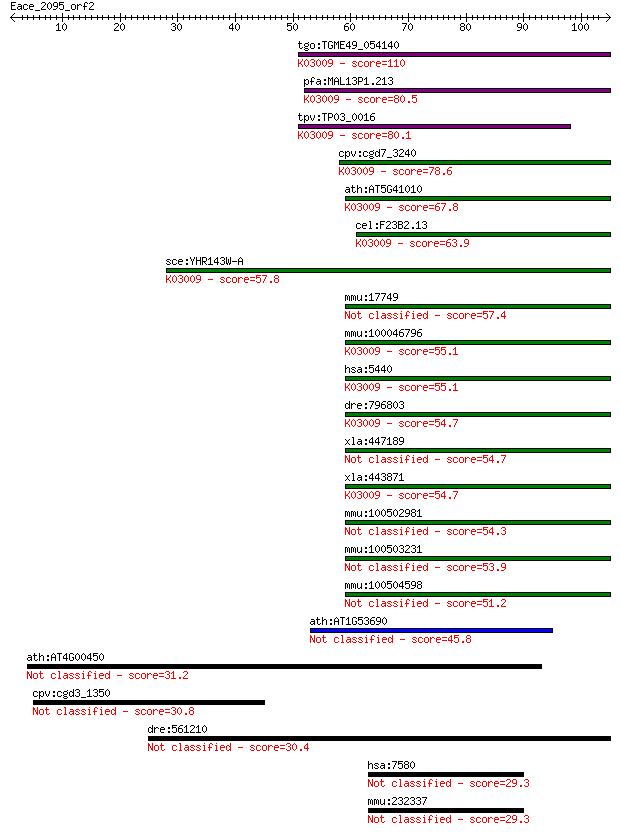
tgo:TGME49_054140 transcription activator, putative ; K03009 D... 110 8e-25
pfa:MAL13P1.213 transcription activator, putative; K03009 DNA-... 80.5 1e-15
tpv:TP03_0016 transcription activator; K03009 DNA-directed RNA... 80.1 2e-15
cpv:cgd7_3240 transcription activator ; K03009 DNA-directed RN... 78.6 5e-15
ath:AT5G41010 NRPB12; NRPB12; DNA binding / DNA-directed RNA p... 67.8 8e-12
cel:F23B2.13 rpb-12; RNA Polymerase II (B) subunit family memb... 63.9 1e-10
sce:YHR143W-A RPC10, RPB12; ABC10-alpha; K03009 DNA-directed R... 57.8 9e-09
mmu:17749 Polr2k, MafY, Mt1a, RPABC4, RPB10alpha, RPB12, RPB7.... 57.4 1e-08
mmu:100046796 DNA-directed RNA polymerases I, II, and III subu... 55.1 5e-08
hsa:5440 POLR2K, ABC10-alpha, RPABC4, RPB10alpha, RPB12, RPB7.... 55.1 5e-08
dre:796803 MGC171795; zgc:171795; K03009 DNA-directed RNA poly... 54.7 7e-08
xla:447189 polr2k-b, abc10-alpha, polr2kb, rpabc4, rpb10alpha,... 54.7 7e-08
xla:443871 polr2k-a, abc10-alpha, polr2ka, rpabc4, rpb10alpha,... 54.7 7e-08
mmu:100502981 DNA-directed RNA polymerases I, II, and III subu... 54.3 1e-07
mmu:100503231 DNA-directed RNA polymerases I, II, and III subu... 53.9 1e-07
mmu:100504598 DNA-directed RNA polymerases I, II, and III subu... 51.2 8e-07
ath:AT1G53690 DNA-directed RNA polymerases I, II, and III 7 kD... 45.8 3e-05
ath:AT4G00450 CRP; CRP (CRYPTIC PRECOCIOUS) 31.2 0.77
cpv:cgd3_1350 UB domain containing protein 30.8 1.0
dre:561210 MGC162148, cbara1; zgc:162148 30.4 1.4
hsa:7580 ZNF32, KOX30; zinc finger protein 32 29.3
mmu:232337 Zfp637, AI646709, BC021601, MGC54806, Znf32; zinc f... 29.3 3.6
> tgo:TGME49_054140 transcription activator, putative ; K03009
DNA-directed RNA polymerases I, II, and III subunit RPABC4
Length=104
Score = 110 bits (276), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 47/54 (87%), Positives = 51/54 (94%), Gaps = 0/54 (0%)
Query 51 IEDNNTQLEPVTYICGECGCDVTLQPTAAVRCRSCGSRILYKKRSRKMMQYEAR 104
++D N+Q+EPVTYICGECGCDV LQP AAVRCRSCGSRILYKKRSRKMMQYEAR
Sbjct 51 LDDGNSQVEPVTYICGECGCDVMLQPNAAVRCRSCGSRILYKKRSRKMMQYEAR 104
> pfa:MAL13P1.213 transcription activator, putative; K03009 DNA-directed
RNA polymerases I, II, and III subunit RPABC4
Length=58
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 31/53 (58%), Positives = 42/53 (79%), Gaps = 0/53 (0%)
Query 52 EDNNTQLEPVTYICGECGCDVTLQPTAAVRCRSCGSRILYKKRSRKMMQYEAR 104
+D + EPV YICGECG D + P A++RC++CGS+I +KKRSR++MQYEAR
Sbjct 6 QDEDISTEPVVYICGECGIDTVIPPNASLRCKNCGSKIFFKKRSRRVMQYEAR 58
> tpv:TP03_0016 transcription activator; K03009 DNA-directed RNA
polymerases I, II, and III subunit RPABC4
Length=58
Score = 80.1 bits (196), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 32/47 (68%), Positives = 39/47 (82%), Gaps = 0/47 (0%)
Query 51 IEDNNTQLEPVTYICGECGCDVTLQPTAAVRCRSCGSRILYKKRSRK 97
DN+ EP+TYICGECG DV+LQP++ VRCR+CGSRILYKKRS +
Sbjct 5 FHDNDDLTEPITYICGECGHDVSLQPSSVVRCRNCGSRILYKKRSYR 51
> cpv:cgd7_3240 transcription activator ; K03009 DNA-directed
RNA polymerases I, II, and III subunit RPABC4
Length=49
Score = 78.6 bits (192), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 37/48 (77%), Positives = 42/48 (87%), Gaps = 1/48 (2%)
Query 58 LEPVTYICGECGCDVTLQPT-AAVRCRSCGSRILYKKRSRKMMQYEAR 104
+EPVTYICGECG DV+L AAVRCR+CG RILYKKR+RKM+QYEAR
Sbjct 2 VEPVTYICGECGADVSLLSHGAAVRCRTCGCRILYKKRTRKMIQYEAR 49
> ath:AT5G41010 NRPB12; NRPB12; DNA binding / DNA-directed RNA
polymerase; K03009 DNA-directed RNA polymerases I, II, and
III subunit RPABC4
Length=51
Score = 67.8 bits (164), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 28/46 (60%), Positives = 38/46 (82%), Gaps = 0/46 (0%)
Query 59 EPVTYICGECGCDVTLQPTAAVRCRSCGSRILYKKRSRKMMQYEAR 104
EPVTY+CG+CG + TL+ ++CR CG RILYKKR+R+++QYEAR
Sbjct 6 EPVTYVCGDCGQENTLKSGDVIQCRECGYRILYKKRTRRVVQYEAR 51
> cel:F23B2.13 rpb-12; RNA Polymerase II (B) subunit family member
(rpb-12); K03009 DNA-directed RNA polymerases I, II, and
III subunit RPABC4
Length=62
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 26/44 (59%), Positives = 33/44 (75%), Gaps = 0/44 (0%)
Query 61 VTYICGECGCDVTLQPTAAVRCRSCGSRILYKKRSRKMMQYEAR 104
+ YICGEC + ++P A+RCR CG RILYKKR RK+M Y+AR
Sbjct 19 MIYICGECHAENEIKPKDAIRCRECGYRILYKKRCRKLMVYDAR 62
> sce:YHR143W-A RPC10, RPB12; ABC10-alpha; K03009 DNA-directed
RNA polymerases I, II, and III subunit RPABC4
Length=70
Score = 57.8 bits (138), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 33/77 (42%), Positives = 43/77 (55%), Gaps = 15/77 (19%)
Query 28 PTTAGATAAAGTTTAAAAAAKMEIEDNNTQLEPVTYICGECGCDVTLQPTAAVRCRSCGS 87
PT A AAAGT+ A A K YIC EC ++L T AVRC+ CG
Sbjct 9 PTNLDA-AAAGTSQARTATLK--------------YICAECSSKLSLSRTDAVRCKDCGH 53
Query 88 RILYKKRSRKMMQYEAR 104
RIL K R+++++Q+EAR
Sbjct 54 RILLKARTKRLVQFEAR 70
> mmu:17749 Polr2k, MafY, Mt1a, RPABC4, RPB10alpha, RPB12, RPB7.0;
polymerase (RNA) II (DNA directed) polypeptide K (EC:2.7.7.6)
Length=99
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 34/46 (73%), Gaps = 0/46 (0%)
Query 59 EPVTYICGECGCDVTLQPTAAVRCRSCGSRILYKKRSRKMMQYEAR 104
+P+ YICGEC + ++ +RCR CG RI+YKKR+++++ ++AR
Sbjct 54 QPMIYICGECHTENEIKSRDPIRCRECGYRIMYKKRTKRLVVFDAR 99
> mmu:100046796 DNA-directed RNA polymerases I, II, and III subunit
RPABC4-like; K03009 DNA-directed RNA polymerases I, II,
and III subunit RPABC4
Length=58
Score = 55.1 bits (131), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 34/46 (73%), Gaps = 0/46 (0%)
Query 59 EPVTYICGECGCDVTLQPTAAVRCRSCGSRILYKKRSRKMMQYEAR 104
+P+ YICGEC + ++ +RCR CG RI+YKKR+++++ ++AR
Sbjct 13 QPMIYICGECHTENEIKSRDPIRCRECGYRIMYKKRTKRLVVFDAR 58
> hsa:5440 POLR2K, ABC10-alpha, RPABC4, RPB10alpha, RPB12, RPB7.0,
hRPB7.0, hsRPB10a; polymerase (RNA) II (DNA directed) polypeptide
K, 7.0kDa (EC:2.7.7.6); K03009 DNA-directed RNA polymerases
I, II, and III subunit RPABC4
Length=58
Score = 55.1 bits (131), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 34/46 (73%), Gaps = 0/46 (0%)
Query 59 EPVTYICGECGCDVTLQPTAAVRCRSCGSRILYKKRSRKMMQYEAR 104
+P+ YICGEC + ++ +RCR CG RI+YKKR+++++ ++AR
Sbjct 13 QPMIYICGECHTENEIKSRDPIRCRECGYRIMYKKRTKRLVVFDAR 58
> dre:796803 MGC171795; zgc:171795; K03009 DNA-directed RNA polymerases
I, II, and III subunit RPABC4
Length=58
Score = 54.7 bits (130), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 34/46 (73%), Gaps = 0/46 (0%)
Query 59 EPVTYICGECGCDVTLQPTAAVRCRSCGSRILYKKRSRKMMQYEAR 104
+P+ YICGEC + ++ +RCR CG RI+YKKR+++++ ++AR
Sbjct 13 QPMIYICGECHTENEIKARDPIRCRECGYRIMYKKRTKRLVVFDAR 58
> xla:447189 polr2k-b, abc10-alpha, polr2kb, rpabc4, rpb10alpha,
rpb12, rpb7.0; polymerase (RNA) II (DNA directed) polypeptide
K, 7.0kDa
Length=58
Score = 54.7 bits (130), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 34/46 (73%), Gaps = 0/46 (0%)
Query 59 EPVTYICGECGCDVTLQPTAAVRCRSCGSRILYKKRSRKMMQYEAR 104
+P+ YICGEC + ++ +RCR CG RI+YKKR+++++ ++AR
Sbjct 13 QPMIYICGECHTENEIKARDPIRCRECGYRIMYKKRTKRLVVFDAR 58
> xla:443871 polr2k-a, abc10-alpha, polr2ka, rpabc4, rpb10alpha,
rpb12, rpb7.0; polymerase (RNA) II (DNA directed) polypeptide
K, 7.0kDa; K03009 DNA-directed RNA polymerases I, II, and
III subunit RPABC4
Length=58
Score = 54.7 bits (130), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 34/46 (73%), Gaps = 0/46 (0%)
Query 59 EPVTYICGECGCDVTLQPTAAVRCRSCGSRILYKKRSRKMMQYEAR 104
+P+ YICGEC + ++ +RCR CG RI+YKKR+++++ ++AR
Sbjct 13 QPMIYICGECHTENEIKARDPIRCRECGYRIMYKKRTKRLVVFDAR 58
> mmu:100502981 DNA-directed RNA polymerases I, II, and III subunit
RPABC4-like
Length=58
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 34/46 (73%), Gaps = 0/46 (0%)
Query 59 EPVTYICGECGCDVTLQPTAAVRCRSCGSRILYKKRSRKMMQYEAR 104
+P+ YICGEC + ++ ++RCR CG RI+YKKR+++++ + AR
Sbjct 13 QPMIYICGECHTENEIKSRDSIRCRECGYRIMYKKRTKRLVVFYAR 58
> mmu:100503231 DNA-directed RNA polymerases I, II, and III subunit
RPABC4-like
Length=58
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 34/46 (73%), Gaps = 0/46 (0%)
Query 59 EPVTYICGECGCDVTLQPTAAVRCRSCGSRILYKKRSRKMMQYEAR 104
+P+ YICGEC + ++ +RCR CG RI+Y+KR+++++ ++AR
Sbjct 13 QPMIYICGECHTENEIKSRDPIRCRECGYRIMYRKRTKRLVVFDAR 58
> mmu:100504598 DNA-directed RNA polymerases I, II, and III subunit
RPABC4-like
Length=58
Score = 51.2 bits (121), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 33/46 (71%), Gaps = 0/46 (0%)
Query 59 EPVTYICGECGCDVTLQPTAAVRCRSCGSRILYKKRSRKMMQYEAR 104
+P+ ICGEC + ++ +RCR CG RI+YKKR+++++ ++AR
Sbjct 13 QPMIDICGECHTENEIKSRDPIRCRECGYRIMYKKRTKRLVVFDAR 58
> ath:AT1G53690 DNA-directed RNA polymerases I, II, and III 7
kDa subunit, putative
Length=61
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 19/42 (45%), Positives = 26/42 (61%), Gaps = 0/42 (0%)
Query 53 DNNTQLEPVTYICGECGCDVTLQPTAAVRCRSCGSRILYKKR 94
D+ + V Y+CG+CG + L+ +CR CG RILYKKR
Sbjct 9 DDKQPEQLVIYVCGDCGQENILKRGDVFQCRDCGFRILYKKR 50
> ath:AT4G00450 CRP; CRP (CRYPTIC PRECOCIOUS)
Length=2253
Score = 31.2 bits (69), Expect = 0.77, Method: Composition-based stats.
Identities = 26/95 (27%), Positives = 43/95 (45%), Gaps = 8/95 (8%)
Query 4 GLSTSAATVATATAAARGSAAAGTPTTAGATAAAGTTT------AAAAAAKMEIEDNNTQ 57
G ++S + +++ + R G+P+ A ++A T T AA A+ +
Sbjct 1932 GTASSGSEISSNKGSTRKGLRGGSPSLARRSSANTTDTSPPPSPAALRASMSLRLQFLLR 1991
Query 58 LEPVTYICGECGCDVTLQPTAAVRCRSCGSRILYK 92
L PV ICGE T A+ R GSR++Y+
Sbjct 1992 LLPV--ICGEPSFKNTRHALASTIVRLLGSRVVYE 2024
> cpv:cgd3_1350 UB domain containing protein
Length=1023
Score = 30.8 bits (68), Expect = 1.0, Method: Composition-based stats.
Identities = 18/41 (43%), Positives = 26/41 (63%), Gaps = 1/41 (2%)
Query 5 LSTSAATVATATAAARG-SAAAGTPTTAGATAAAGTTTAAA 44
LS S+ TVA+ A A+G SA+A PT G+T ++ T + A
Sbjct 528 LSQSSNTVASVAAPAQGESASARAPTDTGSTPSSNTNVSEA 568
> dre:561210 MGC162148, cbara1; zgc:162148
Length=489
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 25/88 (28%), Positives = 39/88 (44%), Gaps = 8/88 (9%)
Query 25 AGTPTTAGATAAAG---TTTAAAAAAKMEIEDNNTQLEPVTYICGECGCDVTLQPTAAVR 81
A G +A+AG T A A + ++ E+ + EP+ + E D L+ ++
Sbjct 37 AALAGVTGISASAGLMWTRAYAEAGSSVKHEEQMREEEPLKDVAEEAESDGALESSSGED 96
Query 82 CRSCGSRILYKK-----RSRKMMQYEAR 104
GS KK R RK+M+YE R
Sbjct 97 EDEAGSEEKKKKQRIGFRDRKVMEYENR 124
> hsa:7580 ZNF32, KOX30; zinc finger protein 32
Length=273
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 12/27 (44%), Positives = 17/27 (62%), Gaps = 0/27 (0%)
Query 63 YICGECGCDVTLQPTAAVRCRSCGSRI 89
Y+CG+CG T + + AV RSC R+
Sbjct 245 YLCGQCGKSFTQRGSLAVHQRSCSQRL 271
> mmu:232337 Zfp637, AI646709, BC021601, MGC54806, Znf32; zinc
finger protein 637
Length=272
Score = 29.3 bits (64), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 12/27 (44%), Positives = 17/27 (62%), Gaps = 0/27 (0%)
Query 63 YICGECGCDVTLQPTAAVRCRSCGSRI 89
Y+CG+CG T + + AV RSC R+
Sbjct 244 YLCGQCGKSFTQRGSLAVHQRSCSQRL 270
Lambda K H
0.312 0.119 0.335
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2022291452
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40