bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2069_orf1
Length=102
Score E
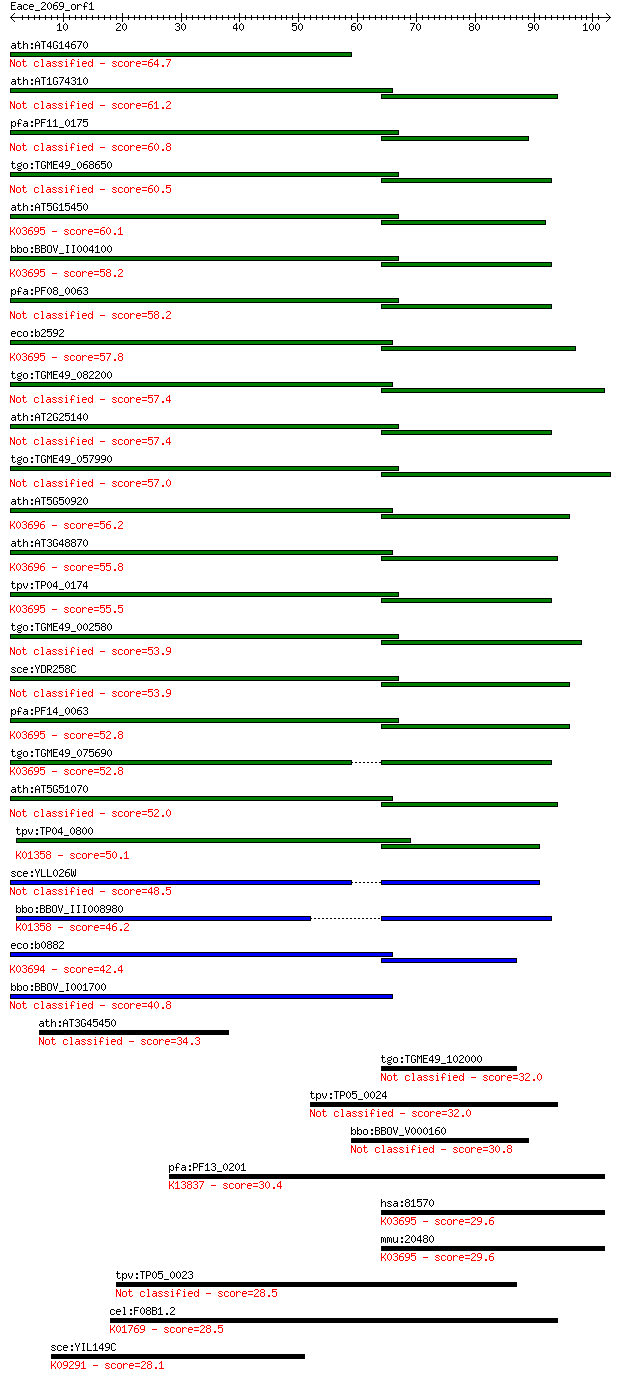
Sequences producing significant alignments: (Bits) Value
ath:AT4G14670 CLPB2; CLPB2; ATP binding / nucleoside-triphosph... 64.7 7e-11
ath:AT1G74310 ATHSP101 (ARABIDOPSIS THALIANA HEAT SHOCK PROTEI... 61.2 7e-10
pfa:PF11_0175 heat shock protein 101, putative 60.8 1e-09
tgo:TGME49_068650 clp ATP-binding chain B1, putative (EC:3.4.2... 60.5 1e-09
ath:AT5G15450 CLPB3; CLPB3 (CASEIN LYTIC PROTEINASE B3); ATP b... 60.1 2e-09
bbo:BBOV_II004100 18.m06340; ClpB; K03695 ATP-dependent Clp pr... 58.2 7e-09
pfa:PF08_0063 ClpB protein, putative 58.2 7e-09
eco:b2592 clpB, ECK2590, htpM, JW2573; protein disaggregation ... 57.8 9e-09
tgo:TGME49_082200 clpB protein, putative 57.4 1e-08
ath:AT2G25140 CLPB4; CLPB4 (CASEIN LYTIC PROTEINASE B4); ATP b... 57.4 1e-08
tgo:TGME49_057990 heat shock protein, putative (EC:3.4.21.53) 57.0 1e-08
ath:AT5G50920 CLPC1; CLPC1; ATP binding / ATP-dependent peptid... 56.2 3e-08
ath:AT3G48870 HSP93-III; ATP binding / ATPase/ DNA binding / n... 55.8 3e-08
tpv:TP04_0174 hypothetical protein; K03695 ATP-dependent Clp p... 55.5 4e-08
tgo:TGME49_002580 heat shock protein, putative (EC:3.4.21.53) 53.9 1e-07
sce:YDR258C HSP78; Hsp78p 53.9 1e-07
pfa:PF14_0063 ATP-dependent CLP protease, putative; K03695 ATP... 52.8 2e-07
tgo:TGME49_075690 chaperone clpB 1 protein, putative (EC:3.4.2... 52.8 3e-07
ath:AT5G51070 ERD1; ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1); ... 52.0 5e-07
tpv:TP04_0800 ATP-dependent Clp protease ATP-binding subunit (... 50.1 2e-06
sce:YLL026W HSP104; Heat shock protein that cooperates with Yd... 48.5 5e-06
bbo:BBOV_III008980 17.m07783; Clp amino terminal domain contai... 46.2 2e-05
eco:b0882 clpA, ECK0873, JW0866, lopD; ATPase and specificity ... 42.4 4e-04
bbo:BBOV_I001700 19.m02115; chaperone clpB 40.8 0.001
ath:AT3G45450 Clp amino terminal domain-containing protein 34.3 0.093
tgo:TGME49_102000 chaperone clpB protein, putative 32.0 0.43
tpv:TP05_0024 clpC; molecular chaperone 32.0 0.46
bbo:BBOV_V000160 clpC 30.8 1.1
pfa:PF13_0201 TRAP, SSP-2, SSP2; Thrombospondin-related anonym... 30.4 1.5
hsa:81570 CLPB, FLJ13152, HSP78, SKD3; ClpB caseinolytic pepti... 29.6 2.3
mmu:20480 Clpb, AL118244, Skd3; ClpB caseinolytic peptidase B ... 29.6 2.7
tpv:TP05_0023 clpC; molecular chaperone 28.5 4.8
cel:F08B1.2 gcy-12; Guanylyl CYclase family member (gcy-12); K... 28.5 5.5
sce:YIL149C MLP2; Mlp2p; K09291 nucleoprotein TPR 28.1 6.7
> ath:AT4G14670 CLPB2; CLPB2; ATP binding / nucleoside-triphosphatase/
nucleotide binding / protein binding
Length=623
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 30/58 (51%), Positives = 38/58 (65%), Gaps = 0/58 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVL 58
ATTLEEY+ H+EKDAAF RRFQ + V PS +SIL+ +K YE + V + D L
Sbjct 279 ATTLEEYRTHVEKDAAFERRFQQVFVAEPSVPDTISILRGLKEKYEGHHGVRIQDRAL 336
> ath:AT1G74310 ATHSP101 (ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN
101); ATP binding / ATPase/ nucleoside-triphosphatase/ nucleotide
binding / protein binding
Length=911
Score = 61.2 bits (147), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 31/65 (47%), Positives = 40/65 (61%), Gaps = 0/65 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATTLEEY+ ++EKDAAF RRFQ + V PS +SIL+ +K YE + V + D L
Sbjct 314 ATTLEEYRKYVEKDAAFERRFQQVYVAEPSVPDTISILRGLKEKYEGHHGVRIQDRALIN 373
Query 61 IVALS 65
LS
Sbjct 374 AAQLS 378
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 16/30 (53%), Positives = 23/30 (76%), Gaps = 0/30 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILEG 93
L+D +G VDF N +II+TSN+G +H+L G
Sbjct 700 LTDGQGRTVDFRNSVIIMTSNLGAEHLLAG 729
> pfa:PF11_0175 heat shock protein 101, putative
Length=906
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 28/66 (42%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATT+ EY+ IE +AF RRF+ I+VE PS + + IL+ +K+ YE FY ++++D+ L
Sbjct 342 ATTIAEYRKFIESCSAFERRFEKILVEPPSVDMTVKILRSLKSKYENFYGINITDKALVA 401
Query 61 IVALSD 66
+SD
Sbjct 402 AAKISD 407
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 10/25 (40%), Positives = 19/25 (76%), Gaps = 0/25 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQ 88
++D+ +DF+N +II+TSN+G +
Sbjct 732 INDNHRRNIDFSNTIIIMTSNLGAE 756
> tgo:TGME49_068650 clp ATP-binding chain B1, putative (EC:3.4.21.53)
Length=929
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 42/66 (63%), Gaps = 0/66 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATTL+EY+ +IEKD A RRFQ ++V+ P E ALSIL+ +K YE + V + D L
Sbjct 331 ATTLDEYRKYIEKDKALERRFQVVLVDEPRVEDALSILRGLKERYEMHHGVSIRDSALVA 390
Query 61 IVALSD 66
LS+
Sbjct 391 ACVLSN 396
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 15/29 (51%), Positives = 21/29 (72%), Gaps = 0/29 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILE 92
L+DS G+ V F NC+II TSN+G +L+
Sbjct 725 LTDSHGHTVSFKNCIIIFTSNMGSDLLLQ 753
> ath:AT5G15450 CLPB3; CLPB3 (CASEIN LYTIC PROTEINASE B3); ATP
binding / ATPase/ nucleoside-triphosphatase/ nucleotide binding
/ protein binding; K03695 ATP-dependent Clp protease ATP-binding
subunit ClpB
Length=968
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/66 (45%), Positives = 42/66 (63%), Gaps = 0/66 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATTL+EY+ +IEKD A RRFQ + V+ P+ E +SIL+ ++ YE + V +SD L
Sbjct 389 ATTLDEYRKYIEKDPALERRFQQVYVDQPTVEDTISILRGLRERYELHHGVRISDSALVE 448
Query 61 IVALSD 66
LSD
Sbjct 449 AAILSD 454
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 16/28 (57%), Positives = 22/28 (78%), Gaps = 0/28 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHIL 91
++DS+G V FTN +II+TSN+G Q IL
Sbjct 779 VTDSQGRTVSFTNTVIIMTSNVGSQFIL 806
> bbo:BBOV_II004100 18.m06340; ClpB; K03695 ATP-dependent Clp
protease ATP-binding subunit ClpB
Length=931
Score = 58.2 bits (139), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 30/66 (45%), Positives = 40/66 (60%), Gaps = 0/66 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATTL+EY+ IEKD A RRFQ + V+ PS E +SIL+ ++ YE + V + D L
Sbjct 355 ATTLQEYRQRIEKDKALERRFQPVYVDQPSVEETISILRGLRERYEVHHGVRILDSALVE 414
Query 61 IVALSD 66
LSD
Sbjct 415 AAQLSD 420
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 19/29 (65%), Positives = 24/29 (82%), Gaps = 0/29 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILE 92
L+DS G +V+FTNC+II TSN+G Q ILE
Sbjct 770 LTDSSGRKVNFTNCMIIFTSNLGSQSILE 798
> pfa:PF08_0063 ClpB protein, putative
Length=1070
Score = 58.2 bits (139), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 40/66 (60%), Gaps = 0/66 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATT+ EY+ IEKD A RRFQ I+VE PS + +SIL+ +K YE + V + D L
Sbjct 467 ATTVSEYRQFIEKDKALERRFQQILVEQPSVDETISILRGLKERYEVHHGVRILDSALVQ 526
Query 61 IVALSD 66
LSD
Sbjct 527 AAVLSD 532
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 17/29 (58%), Positives = 22/29 (75%), Gaps = 0/29 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILE 92
LSD+KGN +F N +II TSN+G Q IL+
Sbjct 910 LSDTKGNVANFRNTIIIFTSNLGSQSILD 938
> eco:b2592 clpB, ECK2590, htpM, JW2573; protein disaggregation
chaperone; K03695 ATP-dependent Clp protease ATP-binding subunit
ClpB
Length=857
Score = 57.8 bits (138), Expect = 9e-09, Method: Composition-based stats.
Identities = 29/65 (44%), Positives = 41/65 (63%), Gaps = 0/65 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATTL+EY+ +IEKDAA RRFQ + V PS E ++IL+ +K YE + V ++D +
Sbjct 313 ATTLDEYRQYIEKDAALERRFQKVFVAEPSVEDTIAILRGLKERYELHHHVQITDPAIVA 372
Query 61 IVALS 65
LS
Sbjct 373 AATLS 377
Score = 33.9 bits (76), Expect = 0.14, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 22/33 (66%), Gaps = 0/33 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILEGYKE 96
L+D +G VDF N ++I+TSN+G I E + E
Sbjct 699 LTDGQGRTVDFRNTVVIMTSNLGSDLIQERFGE 731
> tgo:TGME49_082200 clpB protein, putative
Length=970
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 31/65 (47%), Positives = 43/65 (66%), Gaps = 1/65 (1%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATT EEYK+ IEKDAA RR + I +E PS +RA+ IL+K+ +E + + +SDE +
Sbjct 389 ATTQEEYKI-IEKDAAMERRLKPIFIEEPSTDRAIYILRKLSDKFESHHEMKISDEAIVA 447
Query 61 IVALS 65
V LS
Sbjct 448 AVMLS 452
Score = 48.1 bits (113), Expect = 6e-06, Method: Composition-based stats.
Identities = 22/38 (57%), Positives = 29/38 (76%), Gaps = 0/38 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILEGYKEARALS 101
L+D KG VDFTNC+IILTSN+G Q+I+ Y++A S
Sbjct 716 LTDGKGLLVDFTNCVIILTSNVGAQYIISAYEQAEKES 753
> ath:AT2G25140 CLPB4; CLPB4 (CASEIN LYTIC PROTEINASE B4); ATP
binding / ATPase/ nucleoside-triphosphatase/ nucleotide binding
/ protein binding
Length=964
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 40/66 (60%), Gaps = 0/66 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATTL EY+ +IEKD A RRFQ ++ PS E +SIL+ ++ YE + V +SD L
Sbjct 394 ATTLTEYRKYIEKDPALERRFQQVLCVQPSVEDTISILRGLRERYELHHGVTISDSALVS 453
Query 61 IVALSD 66
L+D
Sbjct 454 AAVLAD 459
Score = 44.7 bits (104), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 17/29 (58%), Positives = 23/29 (79%), Gaps = 0/29 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILE 92
++DS+G V F NC++I+TSNIG HILE
Sbjct 784 ITDSQGRTVSFKNCVVIMTSNIGSHHILE 812
> tgo:TGME49_057990 heat shock protein, putative (EC:3.4.21.53)
Length=921
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 29/66 (43%), Positives = 41/66 (62%), Gaps = 0/66 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATTL+EY+ ++EKDAAF RRFQ + V PS + +SIL+ +K Y + V + D L
Sbjct 311 ATTLDEYRKYVEKDAAFERRFQQVHVREPSVQATISILRGLKDRYASHHGVRILDSALVE 370
Query 61 IVALSD 66
L+D
Sbjct 371 AAQLAD 376
Score = 38.1 bits (87), Expect = 0.008, Method: Composition-based stats.
Identities = 17/39 (43%), Positives = 27/39 (69%), Gaps = 0/39 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILEGYKEARALSA 102
L+DS+G VDF+N +IILTSN+G ++E + ++A
Sbjct 701 LTDSQGRTVDFSNTIIILTSNLGAGFLIEAAQRVDPVAA 739
> ath:AT5G50920 CLPC1; CLPC1; ATP binding / ATP-dependent peptidase/
ATPase; K03696 ATP-dependent Clp protease ATP-binding
subunit ClpC
Length=929
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 28/65 (43%), Positives = 39/65 (60%), Gaps = 0/65 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATTL+EY+ HIEKD A RRFQ + V P+ + + ILK ++ YE + + +DE L
Sbjct 408 ATTLDEYRKHIEKDPALERRFQPVKVPEPTVDETIQILKGLRERYEIHHKLRYTDESLVA 467
Query 61 IVALS 65
LS
Sbjct 468 AAQLS 472
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 17/32 (53%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILEGYK 95
L+DSKG VDF N L+I+TSN+G I +G +
Sbjct 739 LTDSKGRTVDFKNTLLIMTSNVGSSVIEKGGR 770
> ath:AT3G48870 HSP93-III; ATP binding / ATPase/ DNA binding /
nuclease/ nucleoside-triphosphatase/ nucleotide binding / protein
binding; K03696 ATP-dependent Clp protease ATP-binding
subunit ClpC
Length=952
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 28/65 (43%), Positives = 40/65 (61%), Gaps = 0/65 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATT++EY+ HIEKD A RRFQ + V P+ E A+ IL+ ++ YE + + +DE L
Sbjct 429 ATTIDEYRKHIEKDPALERRFQPVKVPEPTVEEAIQILQGLRERYEIHHKLRYTDEALVA 488
Query 61 IVALS 65
LS
Sbjct 489 AAQLS 493
Score = 37.0 bits (84), Expect = 0.015, Method: Composition-based stats.
Identities = 17/30 (56%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILEG 93
L+DSKG VDF N L+I+TSN+G I +G
Sbjct 760 LTDSKGRTVDFKNTLLIMTSNVGSSVIEKG 789
> tpv:TP04_0174 hypothetical protein; K03695 ATP-dependent Clp
protease ATP-binding subunit ClpB
Length=985
Score = 55.5 bits (132), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 41/66 (62%), Gaps = 0/66 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATTL+EY+ IEKD A RRFQ I ++ P+ E ++IL+ +K YE + V + D L
Sbjct 415 ATTLQEYRQKIEKDKALERRFQPIYIDEPNIEETINILRGLKERYEVHHGVRILDSTLIQ 474
Query 61 IVALSD 66
V LS+
Sbjct 475 AVLLSN 480
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 19/29 (65%), Positives = 23/29 (79%), Gaps = 0/29 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILE 92
L+DS G +V+FTN LII TSN+G Q ILE
Sbjct 825 LTDSLGRKVNFTNSLIIFTSNLGSQSILE 853
> tgo:TGME49_002580 heat shock protein, putative (EC:3.4.21.53)
Length=983
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 25/66 (37%), Positives = 39/66 (59%), Gaps = 0/66 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATT EY+ HIE+D AF RRF I ++ P + +++LK ++ N E + + ++D L
Sbjct 414 ATTRAEYRKHIERDMAFARRFVTIEMKEPDVAKTITMLKGIRKNLENHHKLTITDGALVA 473
Query 61 IVALSD 66
LSD
Sbjct 474 AATLSD 479
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 20/34 (58%), Positives = 26/34 (76%), Gaps = 0/34 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILEGYKEA 97
L+D +G VDFTNC+II TSNIG +HIL ++A
Sbjct 729 LTDMRGITVDFTNCVIIATSNIGAKHILAASEKA 762
> sce:YDR258C HSP78; Hsp78p
Length=811
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 41/66 (62%), Gaps = 1/66 (1%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATTL+E+K+ IEKD A RRFQ I++ PS +SIL+ +K YE + V ++D L
Sbjct 249 ATTLDEFKI-IEKDPALSRRFQPILLNEPSVSDTISILRGLKERYEVHHGVRITDTALVS 307
Query 61 IVALSD 66
LS+
Sbjct 308 AAVLSN 313
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 17/32 (53%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILEGYK 95
L+DS G+ VDF N +I++TSNIGQ +L K
Sbjct 635 LTDSLGHHVDFRNTIIVMTSNIGQDILLNDTK 666
> pfa:PF14_0063 ATP-dependent CLP protease, putative; K03695 ATP-dependent
Clp protease ATP-binding subunit ClpB
Length=1341
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 41/66 (62%), Gaps = 0/66 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
TT +EY IE D A RRF + + + + +LKK+K NYEK++++ +D+ L+
Sbjct 657 TTTFQEYSKFIENDKALRRRFNCVTINPFTSKETYKLLKKIKYNYEKYHNIYYTDDSLKS 716
Query 61 IVALSD 66
IV+L++
Sbjct 717 IVSLTE 722
Score = 36.2 bits (82), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILEGYK 95
L+DSKGN+V F N I +T+N+G I + +K
Sbjct 1061 LTDSKGNKVSFKNTFIFMTTNVGSDIITDYFK 1092
> tgo:TGME49_075690 chaperone clpB 1 protein, putative (EC:3.4.21.53);
K03695 ATP-dependent Clp protease ATP-binding subunit
ClpB
Length=898
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 26/58 (44%), Positives = 35/58 (60%), Gaps = 0/58 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVL 58
ATT EY+ +IEKD A RRFQ ++VE P +SIL+ +K YE + V + D L
Sbjct 462 ATTTNEYRQYIEKDKALERRFQKVLVEEPQVSETISILRGLKDRYEVHHGVRILDSAL 519
Score = 45.1 bits (105), Expect = 6e-05, Method: Composition-based stats.
Identities = 18/29 (62%), Positives = 24/29 (82%), Gaps = 0/29 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILE 92
++D KGN V+F NC++I TSN+G QHILE
Sbjct 857 VTDGKGNVVNFRNCIVIFTSNLGSQHILE 885
> ath:AT5G51070 ERD1; ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1);
ATP binding / ATPase/ nucleoside-triphosphatase/ nucleotide
binding / protein binding
Length=945
Score = 52.0 bits (123), Expect = 5e-07, Method: Composition-based stats.
Identities = 24/65 (36%), Positives = 40/65 (61%), Gaps = 0/65 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
+TTL+E++ EKD A RRFQ +++ PS+E A+ IL ++ YE ++ + E ++
Sbjct 427 STTLDEFRSQFEKDKALARRFQPVLINEPSEEDAVKILLGLREKYEAHHNCKYTMEAIDA 486
Query 61 IVALS 65
V LS
Sbjct 487 AVYLS 491
Score = 37.7 bits (86), Expect = 0.008, Method: Composition-based stats.
Identities = 17/30 (56%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILEG 93
L+DS+G RV F N LII+TSN+G I +G
Sbjct 758 LTDSQGRRVSFKNALIIMTSNVGSLAIAKG 787
> tpv:TP04_0800 ATP-dependent Clp protease ATP-binding subunit
(EC:3.4.21.92); K01358 ATP-dependent Clp protease, protease
subunit [EC:3.4.21.92]
Length=900
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 41/67 (61%), Gaps = 2/67 (2%)
Query 2 TTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEGI 61
TT +EYK + EKD A RRF I V+ PS E L IL + ++Y +F+ V+ + + ++
Sbjct 402 TTPKEYKKYFEKDMALSRRFHPIYVDEPSDEDTLKILNGISSSYGEFHGVEYTQDSIK-- 459
Query 62 VALSDSK 68
+AL SK
Sbjct 460 LALKYSK 466
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 17/27 (62%), Gaps = 0/27 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHI 90
L+DSK + F N +II+TSN G I
Sbjct 703 LTDSKNQTISFKNTIIIMTSNTGSNVI 729
> sce:YLL026W HSP104; Heat shock protein that cooperates with
Ydj1p (Hsp40) and Ssa1p (Hsp70) to refold and reactivate previously
denatured, aggregated proteins; responsive to stresses
including: heat, ethanol, and sodium arsenite; involved in
[PSI+] propagation
Length=908
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 25/58 (43%), Positives = 35/58 (60%), Gaps = 0/58 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVL 58
ATT EY+ +EKD AF RRFQ I V PS + ++IL+ ++ YE + V + D L
Sbjct 315 ATTNNEYRSIVEKDGAFERRFQKIEVAEPSVRQTVAILRGLQPKYEIHHGVRILDSAL 372
Score = 31.6 bits (70), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 10/27 (37%), Positives = 20/27 (74%), Gaps = 0/27 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHI 90
++ +G +D +NC++I+TSN+G + I
Sbjct 708 ITSGQGKTIDCSNCIVIMTSNLGAEFI 734
> bbo:BBOV_III008980 17.m07783; Clp amino terminal domain containing
protein; K01358 ATP-dependent Clp protease, protease
subunit [EC:3.4.21.92]
Length=1005
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 23/50 (46%), Positives = 30/50 (60%), Gaps = 0/50 (0%)
Query 2 TTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSV 51
TT +EY+ H EKDAA CRRFQ I V+ PS + IL +F++V
Sbjct 417 TTPKEYQKHFEKDAALCRRFQPIHVKEPSDKDTQIILNATAEACGRFHNV 466
Score = 34.7 bits (78), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 16/29 (55%), Positives = 21/29 (72%), Gaps = 1/29 (3%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILE 92
L+DSK V F N +II+TSN+G HI+E
Sbjct 789 LTDSKNQTVSFKNTIIIMTSNVG-SHIIE 816
> eco:b0882 clpA, ECK0873, JW0866, lopD; ATPase and specificity
subunit of ClpA-ClpP ATP-dependent serine protease, chaperone
activity; K03694 ATP-dependent Clp protease ATP-binding
subunit ClpA
Length=758
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 34/65 (52%), Gaps = 0/65 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
+TT +E+ EKD A RRFQ I + PS E + I+ +K YE + V + + +
Sbjct 321 STTYQEFSNIFEKDRALARRFQKIDITEPSIEETVQIINGLKPKYEAHHDVRYTAKAVRA 380
Query 61 IVALS 65
V L+
Sbjct 381 AVELA 385
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 9/23 (39%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIG 86
L+D+ G + DF N ++++T+N G
Sbjct 586 LTDNNGRKADFRNVVLVMTTNAG 608
> bbo:BBOV_I001700 19.m02115; chaperone clpB
Length=833
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 37/65 (56%), Gaps = 0/65 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
+TT +EY + +D AF RRF+ + + S + L+IL + + E ++ V ++D+ L
Sbjct 305 STTAKEYHQYFRRDRAFERRFEILRLHENSADETLAILHGSRPSLEDYHGVKITDDALVA 364
Query 61 IVALS 65
V LS
Sbjct 365 SVELS 369
> ath:AT3G45450 Clp amino terminal domain-containing protein
Length=341
Score = 34.3 bits (77), Expect = 0.093, Method: Composition-based stats.
Identities = 15/32 (46%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 6 EYKLHIEKDAAFCRRFQNIVVEAPSKERALSI 37
+Y+ HIE D A RRFQ + V P+ E A+ I
Sbjct 278 QYRKHIENDPALERRFQPVKVPEPTVEEAIQI 309
> tgo:TGME49_102000 chaperone clpB protein, putative
Length=240
Score = 32.0 bits (71), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 14/23 (60%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIG 86
L+DS G VDF+N LI TSN+G
Sbjct 66 LTDSLGKCVDFSNTLIFFTSNLG 88
> tpv:TP05_0024 clpC; molecular chaperone
Length=529
Score = 32.0 bits (71), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 28/43 (65%), Gaps = 3/43 (6%)
Query 52 DLSDEVLE-GIVALSDSKGNRVDFTNCLIILTSNIGQQHILEG 93
DL ++L+ G + LS+ G+ ++F N II TSNIG + L+G
Sbjct 361 DLMLQILDKGKLTLSN--GDIINFNNSFIIFTSNIGYTYQLKG 401
> bbo:BBOV_V000160 clpC
Length=551
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 19/30 (63%), Gaps = 2/30 (6%)
Query 59 EGIVALSDSKGNRVDFTNCLIILTSNIGQQ 88
EG+ L+DSKGN F LI TSN+G +
Sbjct 401 EGL--LTDSKGNSCKFNKSLIFFTSNLGSK 428
> pfa:PF13_0201 TRAP, SSP-2, SSP2; Thrombospondin-related anonymous
protein, TRAP; K13837 thrombospondin-related anonymous
protein
Length=574
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 23/74 (31%), Positives = 41/74 (55%), Gaps = 2/74 (2%)
Query 28 APSKERALSILKKVKTNYEKFYSVDLSDEVLEGIVALSDSKGNRVDFTNCLIILTSNIGQ 87
+ +KE+AL I+K + + + +L+D +L+ L+D + NR + ++ILT I
Sbjct 108 SKNKEKALIIIKSLLSTNLPYGKTNLTDALLQVRKHLND-RINRENANQLVVILTDGI-P 165
Query 88 QHILEGYKEARALS 101
I + KE+R LS
Sbjct 166 DSIQDSLKESRKLS 179
> hsa:81570 CLPB, FLJ13152, HSP78, SKD3; ClpB caseinolytic peptidase
B homolog (E. coli); K03695 ATP-dependent Clp protease
ATP-binding subunit ClpB
Length=707
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 24/42 (57%), Gaps = 4/42 (9%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQ----QHILEGYKEARALS 101
L+D KG +D + + I+TSN+ QH L+ +EA +S
Sbjct 476 LTDGKGKTIDCKDAIFIMTSNVASDEIAQHALQLRQEALEMS 517
> mmu:20480 Clpb, AL118244, Skd3; ClpB caseinolytic peptidase
B homolog (E. coli); K03695 ATP-dependent Clp protease ATP-binding
subunit ClpB
Length=677
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 24/42 (57%), Gaps = 4/42 (9%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQ----QHILEGYKEARALS 101
L+D KG +D + + I+TSN+ QH L+ +EA +S
Sbjct 446 LTDGKGKTIDCKDAIFIMTSNVASDEIAQHALQLRQEALEMS 487
> tpv:TP05_0023 clpC; molecular chaperone
Length=502
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 40/69 (57%), Gaps = 5/69 (7%)
Query 19 RRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVL-EGIVALSDSKGNRVDFTNC 77
++ + +V E K ++ +L +++ Y++ + L ++L EGI L DS G +FTN
Sbjct 320 KKSKTLVDEIIDKPNSVVLLDEIEKAYKRLCYIFL--QILDEGI--LIDSSGTVGNFTNS 375
Query 78 LIILTSNIG 86
++ TSN+G
Sbjct 376 FVVFTSNLG 384
> cel:F08B1.2 gcy-12; Guanylyl CYclase family member (gcy-12);
K01769 guanylate cyclase, other [EC:4.6.1.2]
Length=1280
Score = 28.5 bits (62), Expect = 5.5, Method: Composition-based stats.
Identities = 23/85 (27%), Positives = 40/85 (47%), Gaps = 10/85 (11%)
Query 18 CRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEGIVALSDSK----GNRVD 73
C+ +V E SK IL+ + Y D++++G+V + DS+ GN +
Sbjct 735 CQYSVTVVREYCSKGSLHDILRNENLKLDHMYVASFVDDLVKGMVYIHDSELKMHGN-LK 793
Query 74 FTNCLI-----ILTSNIGQQHILEG 93
TNCLI + ++ G + + EG
Sbjct 794 STNCLITSRWTLQIADFGLRELREG 818
> sce:YIL149C MLP2; Mlp2p; K09291 nucleoprotein TPR
Length=1679
Score = 28.1 bits (61), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 21/43 (48%), Gaps = 0/43 (0%)
Query 8 KLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYS 50
KL +EK C+R QNIV++ +E A V K +S
Sbjct 323 KLRLEKSKNECQRLQNIVMDCTKEEEATMTTSAVSPTVGKLFS 365
Lambda K H
0.317 0.132 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2031832220
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40