bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2052_orf2
Length=106
Score E
Sequences producing significant alignments: (Bits) Value
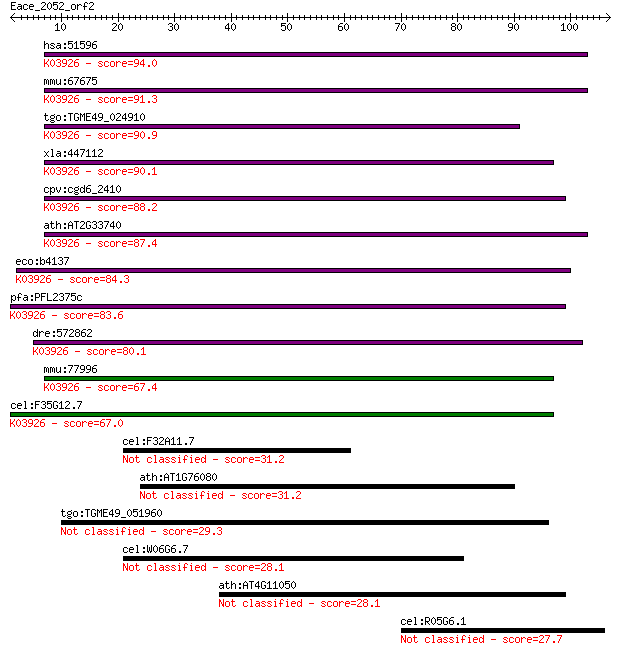
hsa:51596 CUTA, ACHAP, C6orf82, MGC111154; cutA divalent catio... 94.0 1e-19
mmu:67675 Cuta, 0610039D01Rik, 1810022E02Rik, 1810060C03Rik, 2... 91.3 6e-19
tgo:TGME49_024910 CutA1 divalent ion tolerance domain-containi... 90.9 1e-18
xla:447112 cuta, MGC85327, achap; cutA divalent cation toleran... 90.1 1e-18
cpv:cgd6_2410 CutA1 divalent ion tolerance protein ; K03926 pe... 88.2 6e-18
ath:AT2G33740 CUTA; CUTA; copper ion binding; K03926 periplasm... 87.4 1e-17
eco:b4137 cutA, cutA1, cycY, ECK4131, JW4097; divalent-cation ... 84.3 7e-17
pfa:PFL2375c CutA, putative; K03926 periplasmic divalent catio... 83.6 2e-16
dre:572862 MGC174524, MGC63972, cuta; zgc:63972; K03926 peripl... 80.1 2e-15
mmu:77996 D730039F16Rik; RIKEN cDNA D730039F16 gene; K03926 pe... 67.4 1e-11
cel:F35G12.7 hypothetical protein; K03926 periplasmic divalent... 67.0 1e-11
cel:F32A11.7 hypothetical protein 31.2 0.74
ath:AT1G76080 CDSP32; CDSP32 (CHLOROPLASTIC DROUGHT-INDUCED ST... 31.2 0.89
tgo:TGME49_051960 SRS domain-containing protein (EC:3.4.21.53) 29.3 2.8
cel:W06G6.7 hypothetical protein 28.1 6.1
ath:AT4G11050 AtGH9C3 (Arabidopsis thaliana glycosyl hydrolase... 28.1 7.0
cel:R05G6.1 hypothetical protein 27.7 8.8
> hsa:51596 CUTA, ACHAP, C6orf82, MGC111154; cutA divalent cation
tolerance homolog (E. coli); K03926 periplasmic divalent
cation tolerance protein
Length=198
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 43/96 (44%), Positives = 64/96 (66%), Gaps = 0/96 (0%)
Query 7 TAPSNEEARAIAEHLVTQHLAACVQIVPAVESIYEWKGTIEKSSEVLIIIKTQRAHTQAV 66
T P+ + A+ IA +V + LAACV ++P + SIYEWKG IE+ SEVL++IKTQ + A+
Sbjct 93 TCPNEKVAKEIARAVVEKRLAACVNLIPQITSIYEWKGKIEEDSEVLMMIKTQSSLVPAL 152
Query 67 VQAIKDMHSYEVPEVVFTDIVDGNADYIKWARAVTQ 102
++ +H YEV EV+ + GN Y++W R VT+
Sbjct 153 TDFVRSVHPYEVAEVIALPVEQGNFPYLQWVRQVTE 188
> mmu:67675 Cuta, 0610039D01Rik, 1810022E02Rik, 1810060C03Rik,
2700094G22Rik, AI326454; cutA divalent cation tolerance homolog
(E. coli); K03926 periplasmic divalent cation tolerance
protein
Length=177
Score = 91.3 bits (225), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 42/96 (43%), Positives = 63/96 (65%), Gaps = 0/96 (0%)
Query 7 TAPSNEEARAIAEHLVTQHLAACVQIVPAVESIYEWKGTIEKSSEVLIIIKTQRAHTQAV 66
T P+ + A+ IA +V + LAACV ++P + SIYEWKG IE+ SEVL++IKTQ + A+
Sbjct 72 TCPNEKVAKEIARAVVEKRLAACVNLIPQITSIYEWKGKIEEDSEVLMMIKTQSSLVPAL 131
Query 67 VQAIKDMHSYEVPEVVFTDIVDGNADYIKWARAVTQ 102
+ ++ +H YEV EV+ + GN Y+ W VT+
Sbjct 132 TEFVRSVHPYEVAEVIALPVEQGNPPYLHWVHQVTE 167
> tgo:TGME49_024910 CutA1 divalent ion tolerance domain-containing
protein ; K03926 periplasmic divalent cation tolerance
protein
Length=148
Score = 90.9 bits (224), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 41/84 (48%), Positives = 55/84 (65%), Gaps = 0/84 (0%)
Query 7 TAPSNEEARAIAEHLVTQHLAACVQIVPAVESIYEWKGTIEKSSEVLIIIKTQRAHTQAV 66
T +A +A LV LAACV IVP + SIYEW+G +EK EVL+I+KT++ V
Sbjct 22 TCKDKTQAEEVASKLVENRLAACVNIVPGITSIYEWEGKMEKDEEVLLIVKTRKELAAEV 81
Query 67 VQAIKDMHSYEVPEVVFTDIVDGN 90
V A++ HSY+VPEV+F D+ GN
Sbjct 82 VAAVRKWHSYDVPEVIFLDVAGGN 105
> xla:447112 cuta, MGC85327, achap; cutA divalent cation tolerance
homolog; K03926 periplasmic divalent cation tolerance protein
Length=113
Score = 90.1 bits (222), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 40/90 (44%), Positives = 61/90 (67%), Gaps = 0/90 (0%)
Query 7 TAPSNEEARAIAEHLVTQHLAACVQIVPAVESIYEWKGTIEKSSEVLIIIKTQRAHTQAV 66
T P++ A+ IA LV + LAACV ++P + SIYEWKG +E+ +EVL++IKT+ + A+
Sbjct 17 TCPNDTVAKDIARGLVERKLAACVNVIPQITSIYEWKGKLEEDTEVLLMIKTRSSKVSAL 76
Query 67 VQAIKDMHSYEVPEVVFTDIVDGNADYIKW 96
+ ++ +H YEV EV+ I GN Y+KW
Sbjct 77 TEYVRSVHPYEVCEVISLPIEQGNPPYLKW 106
> cpv:cgd6_2410 CutA1 divalent ion tolerance protein ; K03926
periplasmic divalent cation tolerance protein
Length=116
Score = 88.2 bits (217), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 37/92 (40%), Positives = 63/92 (68%), Gaps = 0/92 (0%)
Query 7 TAPSNEEARAIAEHLVTQHLAACVQIVPAVESIYEWKGTIEKSSEVLIIIKTQRAHTQAV 66
+AP+ +EA +IA+ LV + L ACV I+P+V SIY++KG + +EV++++KT +
Sbjct 20 SAPNQDEATSIAKTLVDEELCACVSIIPSVRSIYKFKGQVHDENEVMLLVKTTSQLFTTL 79
Query 67 VQAIKDMHSYEVPEVVFTDIVDGNADYIKWAR 98
+ + ++HSYE+PE++ T +V GN +YI W
Sbjct 80 KEKVTEIHSYELPEIIATKVVYGNENYINWVN 111
> ath:AT2G33740 CUTA; CUTA; copper ion binding; K03926 periplasmic
divalent cation tolerance protein
Length=182
Score = 87.4 bits (215), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 37/96 (38%), Positives = 62/96 (64%), Gaps = 0/96 (0%)
Query 7 TAPSNEEARAIAEHLVTQHLAACVQIVPAVESIYEWKGTIEKSSEVLIIIKTQRAHTQAV 66
T P+ E + +A +V + LAACV IVP +ES+YEW+G ++ SE L+IIKT+++ + +
Sbjct 86 TVPNREAGKKLANSIVQEKLAACVNIVPGIESVYEWEGKVQSDSEELLIIKTRQSLLEPL 145
Query 67 VQAIKDMHSYEVPEVVFTDIVDGNADYIKWARAVTQ 102
+ + H Y+VPEV+ I G+ Y++W + T+
Sbjct 146 TEHVNANHEYDVPEVIALPITGGSDKYLEWLKNSTR 181
> eco:b4137 cutA, cutA1, cycY, ECK4131, JW4097; divalent-cation
tolerance protein, copper sensitivity; K03926 periplasmic
divalent cation tolerance protein
Length=112
Score = 84.3 bits (207), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 36/98 (36%), Positives = 60/98 (61%), Gaps = 0/98 (0%)
Query 2 VVGLSTAPSNEEARAIAEHLVTQHLAACVQIVPAVESIYEWKGTIEKSSEVLIIIKTQRA 61
VV L TAP A+ +A ++ + LAAC ++P S+Y W+G +E+ EV +I+KT +
Sbjct 12 VVVLCTAPDEATAQDLAAKVLAEKLAACATLIPGATSLYYWEGKLEQEYEVQMILKTTVS 71
Query 62 HTQAVVQAIKDMHSYEVPEVVFTDIVDGNADYIKWARA 99
H QA+++ +K H Y+ PE++ + G+ DY+ W A
Sbjct 72 HQQALLECLKSHHPYQTPELLVLPVTHGDTDYLSWLNA 109
> pfa:PFL2375c CutA, putative; K03926 periplasmic divalent cation
tolerance protein
Length=159
Score = 83.6 bits (205), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 38/98 (38%), Positives = 59/98 (60%), Gaps = 0/98 (0%)
Query 1 LVVGLSTAPSNEEARAIAEHLVTQHLAACVQIVPAVESIYEWKGTIEKSSEVLIIIKTQR 60
+V T PS E A I+ L+ + L +CV ++P + S+Y WKG I K +EVL++IKT++
Sbjct 56 FIVVYVTTPSKEVAEKISYVLLEEKLVSCVNVIPGILSLYHWKGEIAKDNEVLMMIKTKK 115
Query 61 AHTQAVVQAIKDMHSYEVPEVVFTDIVDGNADYIKWAR 98
+V+ +K H YE+PEV+ I G+ DY+ W
Sbjct 116 HLFDEIVKLVKSNHPYEIPEVIAVPIEYGSKDYLDWVN 153
> dre:572862 MGC174524, MGC63972, cuta; zgc:63972; K03926 periplasmic
divalent cation tolerance protein
Length=150
Score = 80.1 bits (196), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 36/98 (36%), Positives = 61/98 (62%), Gaps = 1/98 (1%)
Query 5 LSTAPSNEEARAIAEHLVTQHLAACVQIVPAVESIYEWKGTIEKSSEVLIIIKTQRAHTQ 64
L P+ + AR I ++ + LAACV I P ++Y WKG I ++E+L++++T+ + Q
Sbjct 51 LVNCPTEQTARDIGRIIMEKRLAACVNIFPRTATMYYWKGEIRDATEILLLVRTKTSLVQ 110
Query 65 AVVQAIKDMHSYEVPEVVFTDIVDGNADYIKW-ARAVT 101
++ I +H Y++PE++ I DG+ Y+KW A AVT
Sbjct 111 RLMTYITAIHPYDIPEIITFPINDGSQHYLKWIAEAVT 148
> mmu:77996 D730039F16Rik; RIKEN cDNA D730039F16 gene; K03926
periplasmic divalent cation tolerance protein
Length=156
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 53/90 (58%), Gaps = 0/90 (0%)
Query 7 TAPSNEEARAIAEHLVTQHLAACVQIVPAVESIYEWKGTIEKSSEVLIIIKTQRAHTQAV 66
P+ + AR IA ++ + +A+ V I+P S+Y WKG IE+ EV ++IKT+ + +
Sbjct 59 NCPNEQIARDIARAILDKKMASSVNILPKTSSLYFWKGEIEEGIEVSLLIKTKTSKVSRL 118
Query 67 VQAIKDMHSYEVPEVVFTDIVDGNADYIKW 96
++ H +E+PEV + G+A +++W
Sbjct 119 FAYMRLAHPFEIPEVFSIPMDQGDARFLRW 148
> cel:F35G12.7 hypothetical protein; K03926 periplasmic divalent
cation tolerance protein
Length=115
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 35/97 (36%), Positives = 55/97 (56%), Gaps = 2/97 (2%)
Query 1 LVVGLSTAPSNEEARAIAEHLVTQHLAACVQIVPAVESIYEWKGTIEKSSEVLIIIKTQR 60
+VV TAPS E A +A VT+ LAAC ++P V S+Y+W+G IE+ E ++I+KT
Sbjct 8 MVVAYVTAPSKEVAMTVARTTVTEALAACANVIPEVTSVYKWQGKIEEDQEHVVILKTVE 67
Query 61 AHTQAVVQAIKDMHSYEVPEVVFTDIVDG-NADYIKW 96
+ + + ++ +H E P FT +D D+ W
Sbjct 68 SKVEELSARVRSLHPAETP-CFFTLAIDKITPDFGGW 103
> cel:F32A11.7 hypothetical protein
Length=657
Score = 31.2 bits (69), Expect = 0.74, Method: Composition-based stats.
Identities = 13/40 (32%), Positives = 27/40 (67%), Gaps = 0/40 (0%)
Query 21 LVTQHLAACVQIVPAVESIYEWKGTIEKSSEVLIIIKTQR 60
LVT+HL++ V+++ + + +W+ TIE+ ++ IK Q+
Sbjct 141 LVTRHLSSAVKLLKKTDKVSKWQDTIERITKKAEEIKEQK 180
> ath:AT1G76080 CDSP32; CDSP32 (CHLOROPLASTIC DROUGHT-INDUCED
STRESS PROTEIN OF 32 KD)
Length=302
Score = 31.2 bits (69), Expect = 0.89, Method: Compositional matrix adjust.
Identities = 17/66 (25%), Positives = 30/66 (45%), Gaps = 8/66 (12%)
Query 24 QHLAACVQIVPAVESIYEWKGTIEKSSEVLIIIKTQRAHTQAVVQAIKDMHSYEVPEVVF 83
+H CV++ P V + SE ++ + + ++ +KDM+ EVP +F
Sbjct 217 KHCGPCVKVYPTVLKLSR------SMSETVVFARMNGDENDSCMEFLKDMNVIEVPTFLF 270
Query 84 TDIVDG 89
I DG
Sbjct 271 --IRDG 274
> tgo:TGME49_051960 SRS domain-containing protein (EC:3.4.21.53)
Length=853
Score = 29.3 bits (64), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 26/98 (26%), Positives = 47/98 (47%), Gaps = 12/98 (12%)
Query 10 SNEEARAIAEHLVTQHLAACVQIVPAVESIYEWKGTIEKSSEVLIIIKTQRAH----TQA 65
++EEAR ++++ I + I E K I K+ + L+ + Q H +
Sbjct 172 ASEEARKKILETLSRYYDLVRVITLVIAGIIERKVKIIKAIKALLQLANQTVHDLLEREE 231
Query 66 VVQAIKDMHSY---EVPEV-----VFTDIVDGNADYIK 95
VVQAI+D+++Y +PE+ + + G DY+K
Sbjct 232 VVQAIEDLYNYIKQHIPEIDKHLKAIVEAIKGLVDYVK 269
> cel:W06G6.7 hypothetical protein
Length=511
Score = 28.1 bits (61), Expect = 6.1, Method: Composition-based stats.
Identities = 15/60 (25%), Positives = 32/60 (53%), Gaps = 4/60 (6%)
Query 21 LVTQHLAACVQIVPAVESIYEWKGTIEKSSEVLIIIKTQRAHTQAVVQAIKDMHSYEVPE 80
++ +HL+ ++++ + +W+ TI + SE IK Q+ + + I+ YE+PE
Sbjct 6 VLAKHLSHTLRLIKNKRTTSKWEDTISRISEKASQIKQQK----DMERRIRKQRGYELPE 61
> ath:AT4G11050 AtGH9C3 (Arabidopsis thaliana glycosyl hydrolase
9C3); carbohydrate binding / catalytic/ hydrolase, hydrolyzing
O-glycosyl compounds (EC:3.2.1.4)
Length=626
Score = 28.1 bits (61), Expect = 7.0, Method: Composition-based stats.
Identities = 16/61 (26%), Positives = 30/61 (49%), Gaps = 6/61 (9%)
Query 38 SIYEWKGTIEKSSEVLIIIKTQRAHTQAVVQAIKDMHSYEVPEVVFTDIVDGNADYIKWA 97
SI E+ G +E + E+ I + T ++A + P V++ ++ DG +D+ W
Sbjct 97 SIIEYGGQLESNGELGHAIDAVKWGTDYFIKA------HPEPNVLYGEVGDGKSDHYCWQ 150
Query 98 R 98
R
Sbjct 151 R 151
> cel:R05G6.1 hypothetical protein
Length=314
Score = 27.7 bits (60), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 70 IKDMHSYEVPEVVFTDIVDGNADYIKWARAVTQPKG 105
+ M YE+ E+ T ++ N D +K AR +TQ G
Sbjct 194 VSGMTVYELRELGGTILIQTNTDELKGARVMTQTSG 229
Lambda K H
0.316 0.128 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2012750684
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40