bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1816_orf1
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
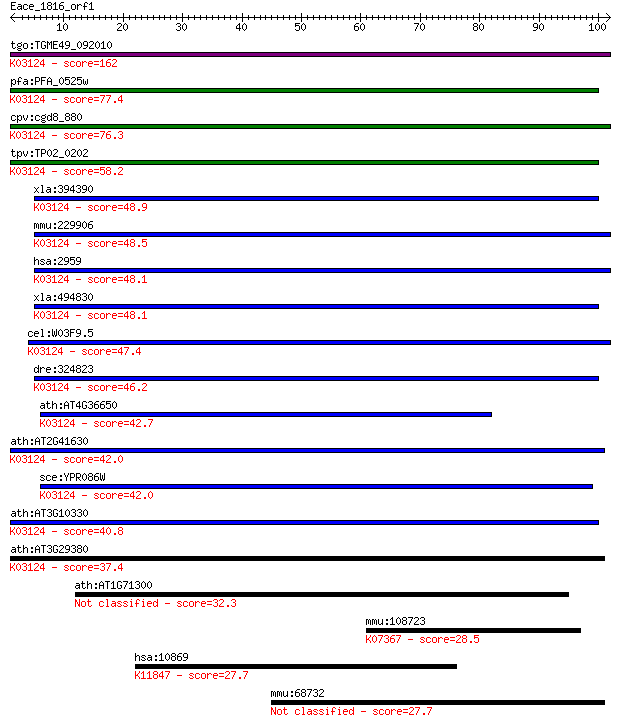
tgo:TGME49_092010 transcription initiation factor IIB, putativ... 162 4e-40
pfa:PFA_0525w transcription initiation factor TFIIB, putative;... 77.4 9e-15
cpv:cgd8_880 transcription initiation factor TFIIB Sua7p; ZnR+... 76.3 2e-14
tpv:TP02_0202 transcription initiation factor TFIIB; K03124 tr... 58.2 6e-09
xla:394390 gtf2b-a, XLTFIIB, gtf2b, tf2b, tfiib; general trans... 48.9 4e-06
mmu:229906 Gtf2b, MGC6859; general transcription factor IIB; K... 48.5 6e-06
hsa:2959 GTF2B, TF2B, TFIIB; general transcription factor IIB;... 48.1 6e-06
xla:494830 gtf2b-b, gtf2b, tf2b, tfiib; general transcription ... 48.1 6e-06
cel:W03F9.5 ttb-1; TFII(two)B (general transcription factor) f... 47.4 1e-05
dre:324823 gtf2b, wu:fc44c11, zgc:66258; general transcription... 46.2 3e-05
ath:AT4G36650 ATPBRP; ATPBRP (PLANT-SPECIFIC TFIIB-RELATED PRO... 42.7 3e-04
ath:AT2G41630 TFIIB; TFIIB (TRANSCRIPTION FACTOR II B); RNA po... 42.0 4e-04
sce:YPR086W SUA7, SOH4; Sua7p; K03124 transcription initiation... 42.0 5e-04
ath:AT3G10330 transcription initiation factor IIB-2 / general ... 40.8 0.001
ath:AT3G29380 transcription factor IIB (TFIIB) family protein;... 37.4 0.012
ath:AT1G71300 Vps52/Sac2 family protein 32.3 0.39
mmu:108723 Card11, 0610008L17Rik, 2410011D02Rik, BIMP3, CARMA1... 28.5 5.0
hsa:10869 USP19, ZMYND9; ubiquitin specific peptidase 19 (EC:3... 27.7 8.8
mmu:68732 Lrrc16a, 1110037D04Rik, AI425970, CARMIL, D130057M20... 27.7 9.8
> tgo:TGME49_092010 transcription initiation factor IIB, putative
; K03124 transcription initiation factor TFIIB
Length=464
Score = 162 bits (409), Expect = 4e-40, Method: Compositional matrix adjust.
Identities = 82/107 (76%), Positives = 90/107 (84%), Gaps = 9/107 (8%)
Query 1 NATHLLPRYCSRLQLSMHISDVAEHVAKRAAQVFVSSH------RPNSVAAAAIWLVVQL 54
+AT LLPRYCSRLQLSMH++DVAEHVAKRA QV +SSH RPNSVAAAAIWLVV+L
Sbjct 333 SATQLLPRYCSRLQLSMHVADVAEHVAKRATQVIISSHRHVLARRPNSVAAAAIWLVVKL 392
Query 55 LSSSSAQQQMLLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLPNE 101
L+SS+ LPK SE+ASVTGAGEHTLRSIYKDMLDVAEHLLP E
Sbjct 393 LNSSTNPN---LPKASEIASVTGAGEHTLRSIYKDMLDVAEHLLPRE 436
> pfa:PFA_0525w transcription initiation factor TFIIB, putative;
K03124 transcription initiation factor TFIIB
Length=367
Score = 77.4 bits (189), Expect = 9e-15, Method: Composition-based stats.
Identities = 35/99 (35%), Positives = 69/99 (69%), Gaps = 2/99 (2%)
Query 1 NATHLLPRYCSRLQLSMHISDVAEHVAKRAAQVFVSSHRPNSVAAAAIWLVVQLLSSSSA 60
N +HL+ +RLQLS+ + + E+V K+A+ + +SHR NS+ +I L+V+L +++
Sbjct 244 NISHLIYSLSNRLQLSIDLIEAIEYVVKKASTLITTSHRLNSLCGGSIHLIVEL--NTNE 301
Query 61 QQQMLLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLP 99
++ M LP +S++A+V G +TL++ +K++L+ AE+++P
Sbjct 302 EKNMKLPNLSQIATVCGVTTNTLKTTFKELLNAAEYIIP 340
> cpv:cgd8_880 transcription initiation factor TFIIB Sua7p; ZnR+2cyclins
; K03124 transcription initiation factor TFIIB
Length=456
Score = 76.3 bits (186), Expect = 2e-14, Method: Composition-based stats.
Identities = 43/101 (42%), Positives = 60/101 (59%), Gaps = 8/101 (7%)
Query 1 NATHLLPRYCSRLQLSMHISDVAEHVAKRAAQVFVSSHRPNSVAAAAIWLVVQLLSSSSA 60
+A L+PR+C LQLS I +AE+V + A Q SHRPNS+AA AI+ V L +
Sbjct 344 SAAELMPRFCHYLQLSHEIVGIAEYVCRTAEQYINKSHRPNSLAAGAIYFVCNL---CNI 400
Query 61 QQQMLLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLPNE 101
Q +M VA +GE T+R +YK++L V E LLP++
Sbjct 401 QIEM-----KSVAVAAKSGETTVRGVYKELLLVGEKLLPSD 436
> tpv:TP02_0202 transcription initiation factor TFIIB; K03124
transcription initiation factor TFIIB
Length=342
Score = 58.2 bits (139), Expect = 6e-09, Method: Composition-based stats.
Identities = 30/99 (30%), Positives = 55/99 (55%), Gaps = 5/99 (5%)
Query 1 NATHLLPRYCSRLQLSMHISDVAEHVAKRAAQVFVSSHRPNSVAAAAIWLVVQLLSSSSA 60
+ + L+PR+CSRL LS I E VA+++ + S+HR S+A I+LV +L + +
Sbjct 210 DVSQLMPRFCSRLNLSNEIMTTCEAVAQKSFVLLTSAHRTTSLAGGIIYLVTRLFFADDS 269
Query 61 QQQMLLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLP 99
+S+++ V G T+++ YK++ + L+P
Sbjct 270 PIS-----ISDISQVCGTSTGTIKTTYKELCLYLDKLIP 303
> xla:394390 gtf2b-a, XLTFIIB, gtf2b, tf2b, tfiib; general transcription
factor 2B; K03124 transcription initiation factor
TFIIB
Length=316
Score = 48.9 bits (115), Expect = 4e-06, Method: Composition-based stats.
Identities = 28/96 (29%), Positives = 47/96 (48%), Gaps = 9/96 (9%)
Query 5 LLPRYCSRLQLSMHISDVAEHVAKRAAQV-FVSSHRPNSVAAAAIWLVVQLLSSSSAQQQ 63
+ R+CS L L+ + A H+A++A ++ V P SVAAAAI++ Q + Q+
Sbjct 214 FMSRFCSNLGLTKQVQMAATHIARKAVELDLVPGRSPISVAAAAIYMASQASAEKRTQK- 272
Query 64 MLLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLP 99
E+ + G + T+R Y+ + A L P
Sbjct 273 -------EIGDIAGVADVTIRQSYRLIYPRAPDLFP 301
> mmu:229906 Gtf2b, MGC6859; general transcription factor IIB;
K03124 transcription initiation factor TFIIB
Length=316
Score = 48.5 bits (114), Expect = 6e-06, Method: Composition-based stats.
Identities = 28/98 (28%), Positives = 48/98 (48%), Gaps = 9/98 (9%)
Query 5 LLPRYCSRLQLSMHISDVAEHVAKRAAQV-FVSSHRPNSVAAAAIWLVVQLLSSSSAQQQ 63
+ R+CS L L + A H+A++A ++ V P SVAAAAI++ Q + Q+
Sbjct 214 FMSRFCSNLCLPKQVQMAATHIARKAVELDLVPGRSPISVAAAAIYMASQASAEKRTQK- 272
Query 64 MLLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLPNE 101
E+ + G + T+R Y+ + A L P++
Sbjct 273 -------EIGDIAGVADVTIRQSYRLIYPRAPDLFPSD 303
> hsa:2959 GTF2B, TF2B, TFIIB; general transcription factor IIB;
K03124 transcription initiation factor TFIIB
Length=316
Score = 48.1 bits (113), Expect = 6e-06, Method: Composition-based stats.
Identities = 28/98 (28%), Positives = 47/98 (47%), Gaps = 9/98 (9%)
Query 5 LLPRYCSRLQLSMHISDVAEHVAKRAAQV-FVSSHRPNSVAAAAIWLVVQLLSSSSAQQQ 63
+ R+CS L L + A H+A++A ++ V P SVAAAAI++ Q + Q+
Sbjct 214 FMSRFCSNLCLPKQVQMAATHIARKAVELDLVPGRSPISVAAAAIYMASQASAEKRTQK- 272
Query 64 MLLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLPNE 101
E+ + G + T+R Y+ + A L P +
Sbjct 273 -------EIGDIAGVADVTIRQSYRLIYPRAPDLFPTD 303
> xla:494830 gtf2b-b, gtf2b, tf2b, tfiib; general transcription
factor 2B; K03124 transcription initiation factor TFIIB
Length=316
Score = 48.1 bits (113), Expect = 6e-06, Method: Composition-based stats.
Identities = 28/96 (29%), Positives = 47/96 (48%), Gaps = 9/96 (9%)
Query 5 LLPRYCSRLQLSMHISDVAEHVAKRAAQV-FVSSHRPNSVAAAAIWLVVQLLSSSSAQQQ 63
+ R+CS L L+ + A H+A++A ++ V P SVAAAAI++ Q + Q+
Sbjct 214 FMSRFCSNLGLTKQVQMAATHIARKAVELDLVPGRSPISVAAAAIYMASQASAEKRTQK- 272
Query 64 MLLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLP 99
E+ + G + T+R Y+ + A L P
Sbjct 273 -------EIGDIAGVADVTIRQSYRLIYPRAPDLFP 301
> cel:W03F9.5 ttb-1; TFII(two)B (general transcription factor)
family member (ttb-1); K03124 transcription initiation factor
TFIIB
Length=306
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 32/99 (32%), Positives = 48/99 (48%), Gaps = 9/99 (9%)
Query 4 HLLPRYCSRLQLSMHISDVAEHVAKRAAQV-FVSSHRPNSVAAAAIWLVVQLLSSSSAQQ 62
+ R+C L L I A +AK A + V+ P S+AAAAI++ Q +SA++
Sbjct 203 DFMSRFCGNLSLPNSIQAAATRIAKCAVDMDLVAGRTPISIAAAAIYMASQ----ASAEK 258
Query 63 QMLLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLPNE 101
+ E+ V GA E T+R YK + A L P +
Sbjct 259 R----SAKEIGDVAGAAEITVRQTYKLLYPKALELFPKD 293
> dre:324823 gtf2b, wu:fc44c11, zgc:66258; general transcription
factor IIB; K03124 transcription initiation factor TFIIB
Length=316
Score = 46.2 bits (108), Expect = 3e-05, Method: Composition-based stats.
Identities = 27/96 (28%), Positives = 46/96 (47%), Gaps = 9/96 (9%)
Query 5 LLPRYCSRLQLSMHISDVAEHVAKRAAQV-FVSSHRPNSVAAAAIWLVVQLLSSSSAQQQ 63
+ R+CS L L + A ++A++A ++ V P SVAAAAI++ Q + Q+
Sbjct 214 FMSRFCSNLGLPKQVQMAATYIARKAVELDLVPGRSPISVAAAAIYMASQASAEKKTQK- 272
Query 64 MLLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLP 99
E+ + G + T+R Y+ + A L P
Sbjct 273 -------EIGDIAGVADVTIRQSYRLIYPRAADLFP 301
> ath:AT4G36650 ATPBRP; ATPBRP (PLANT-SPECIFIC TFIIB-RELATED PROTEIN);
RNA polymerase II transcription factor/ rDNA binding;
K03124 transcription initiation factor TFIIB
Length=369
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 39/77 (50%), Gaps = 9/77 (11%)
Query 6 LPRYCSRLQLSMHISDVAEHVAKRA-AQVFVSSHRPNSVAAAAIWLVVQLLSSSSAQQQM 64
+PR+C+ LQL+ ++A H+ + + F + P S++AAAI+L QL Q
Sbjct 233 MPRFCTLLQLNKSAQELATHIGEVVINKCFCTRRNPISISAAAIYLACQLEDKRKTQ--- 289
Query 65 LLPKVSEVASVTGAGEH 81
+E+ +TG E
Sbjct 290 -----AEICKITGLTER 301
> ath:AT2G41630 TFIIB; TFIIB (TRANSCRIPTION FACTOR II B); RNA
polymerase II transcription factor/ protein binding / transcription
regulator/ translation initiation factor/ zinc ion binding;
K03124 transcription initiation factor TFIIB
Length=312
Score = 42.0 bits (97), Expect = 4e-04, Method: Composition-based stats.
Identities = 25/100 (25%), Positives = 47/100 (47%), Gaps = 9/100 (9%)
Query 1 NATHLLPRYCSRLQLSMHISDVAEHVAKRAAQVFVSSHRPNSVAAAAIWLVVQLLSSSSA 60
+A + R+CS L +S H A+ +++ + F P S+AA I+++ QL
Sbjct 208 HAGDFMRRFCSNLAMSNHAVKAAQEAVQKSEE-FDIRRSPISIAAVVIYIITQLSDDKKT 266
Query 61 QQQMLLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLPN 100
+ +++ TG E T+R+ YKD+ + P+
Sbjct 267 --------LKDISHATGVAEGTIRNSYKDLYPHLSKIAPS 298
> sce:YPR086W SUA7, SOH4; Sua7p; K03124 transcription initiation
factor TFIIB
Length=345
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 27/94 (28%), Positives = 50/94 (53%), Gaps = 9/94 (9%)
Query 6 LPRYCSRLQLSMHISDVAEHVAKRAAQVF-VSSHRPNSVAAAAIWLVVQLLSSSSAQQQM 64
+PR+CS L L M ++ AE+ AK+ ++ ++ P ++A +I+L + L Q +
Sbjct 239 IPRFCSHLGLPMQVTTSAEYTAKKCKEIKEIAGKSPITIAVVSIYLNILLF-----QIPI 293
Query 65 LLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLL 98
KV + VT E T++S YK + + + L+
Sbjct 294 TAAKVGQTLQVT---EGTIKSGYKILYEHRDKLV 324
> ath:AT3G10330 transcription initiation factor IIB-2 / general
transcription factor TFIIB-2 (TFIIB2); K03124 transcription
initiation factor TFIIB
Length=312
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 26/99 (26%), Positives = 51/99 (51%), Gaps = 9/99 (9%)
Query 1 NATHLLPRYCSRLQLSMHISDVAEHVAKRAAQVFVSSHRPNSVAAAAIWLVVQLLSSSSA 60
+A + R+CS L ++ A+ +++ + F P S+AAA I+++ QL S
Sbjct 208 HAGDFMRRFCSNLGMTNQTVKAAQESVQKSEE-FDIRRSPISIAAAVIYIITQL----SD 262
Query 61 QQQMLLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLP 99
+++ L +++ TG E T+R+ YKD+ ++P
Sbjct 263 EKKPL----RDISVATGVAEGTIRNSYKDLYPHLSKIIP 297
> ath:AT3G29380 transcription factor IIB (TFIIB) family protein;
K03124 transcription initiation factor TFIIB
Length=336
Score = 37.4 bits (85), Expect = 0.012, Method: Composition-based stats.
Identities = 30/101 (29%), Positives = 48/101 (47%), Gaps = 10/101 (9%)
Query 1 NATHLLPRYCSRLQLSM-HISDVAEHVAKRAAQVFVSSHRPNSVAAAAIWLVVQLLSSSS 59
N L+ R+CS+L +S I + E V K A+ F P SV AA I+++ +
Sbjct 219 NTGELVRRFCSKLDISQREIMAIPEAVEK--AENFDIRRNPKSVLAAIIFMISHI----- 271
Query 60 AQQQMLLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLPN 100
Q + E+ V E+T+++ KDM A ++PN
Sbjct 272 --SQTNRKPIREIGIVAEVVENTIKNSVKDMYPYALKIIPN 310
> ath:AT1G71300 Vps52/Sac2 family protein
Length=701
Score = 32.3 bits (72), Expect = 0.39, Method: Composition-based stats.
Identities = 24/85 (28%), Positives = 42/85 (49%), Gaps = 4/85 (4%)
Query 12 RLQLSMHISDVAEHVAKRAAQVFVSSHRP--NSVAAAAIWLVVQLLSSSSAQQQMLLPKV 69
+LQL + S+ V R+ +F S P N A A+ +Q++ Q L+P +
Sbjct 296 KLQLDIATSNDLIGVDTRSTGLFSRSKEPLKNRCAVFALGERIQIIKE--IDQPALIPHI 353
Query 70 SEVASVTGAGEHTLRSIYKDMLDVA 94
+E +S+ E RS++K ++D A
Sbjct 354 AEASSLKYPYEVLFRSLHKLLMDTA 378
> mmu:108723 Card11, 0610008L17Rik, 2410011D02Rik, BIMP3, CARMA1;
caspase recruitment domain family, member 11; K07367 caspase
recruitment domain-containing protein 11
Length=1154
Score = 28.5 bits (62), Expect = 5.0, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 61 QQQMLLPKVSEVASVTGAGEHTLRSIYKDMLDVAEH 96
+ +ML K+ E+ S+ AG+ +L K +LD+ EH
Sbjct 272 ENEMLKTKIQELQSIIQAGKRSLPDSDKAILDILEH 307
> hsa:10869 USP19, ZMYND9; ubiquitin specific peptidase 19 (EC:3.4.19.12);
K11847 ubiquitin carboxyl-terminal hydrolase 19
[EC:3.1.2.15]
Length=1419
Score = 27.7 bits (60), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 33/54 (61%), Gaps = 0/54 (0%)
Query 22 VAEHVAKRAAQVFVSSHRPNSVAAAAIWLVVQLLSSSSAQQQMLLPKVSEVASV 75
+AE + R +VF+ SH ++V+ + L +LLSS A++++++ +V + V
Sbjct 831 LAEVIKNRFHRVFLPSHSLDTVSPSDTLLCFELLSSELAKERVVVLEVQQRPQV 884
> mmu:68732 Lrrc16a, 1110037D04Rik, AI425970, CARMIL, D130057M20,
Lrrc16; leucine rich repeat containing 16A
Length=1374
Score = 27.7 bits (60), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 25/56 (44%), Gaps = 4/56 (7%)
Query 45 AAAIWLVVQLLSSSSAQQQMLLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLPN 100
A W LSSS Q+ + S VT + L+ + + M+D AE L PN
Sbjct 745 GGASWAGASGLSSSPIQETL----ESMAGEVTRVVDEQLKDLLESMVDAAETLCPN 796
Lambda K H
0.317 0.125 0.349
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2036602604
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40