bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1693_orf2
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
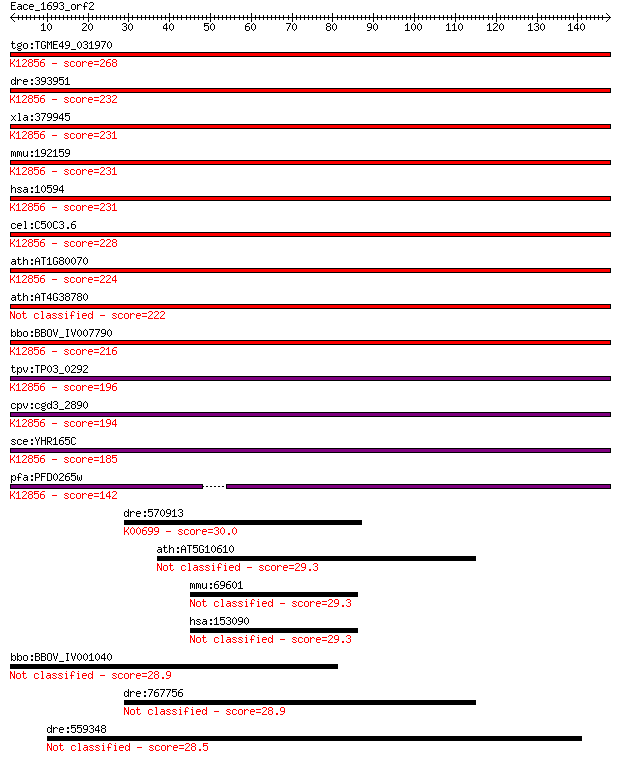
tgo:TGME49_031970 pre-mRNA splicing factor PRP8, putative ; K1... 268 4e-72
dre:393951 prpf8, MGC162871, MGC56504, id:ibd1257, ik:tdsubc_2... 232 4e-61
xla:379945 prpf8, MGC52804, hprp8, prp-8, prp8, prpc8, rp13; P... 231 4e-61
mmu:192159 Prpf8, AU019467, D11Bwg0410e, DBF3/PRP8, Prp8, Sfpr... 231 5e-61
hsa:10594 PRPF8, HPRP8, PRP8, PRPC8, RP13; PRP8 pre-mRNA proce... 231 9e-61
cel:C50C3.6 prp-8; yeast PRP (splicing factor) related family ... 228 5e-60
ath:AT1G80070 SUS2; SUS2 (ABNORMAL SUSPENSOR 2); K12856 pre-mR... 224 7e-59
ath:AT4G38780 splicing factor, putative 222 3e-58
bbo:BBOV_IV007790 23.m06497; processing splicing factor 8; K12... 216 1e-56
tpv:TP03_0292 splicing factor Prp8; K12856 pre-mRNA-processing... 196 2e-50
cpv:cgd3_2890 Prp8. JAB/PAD domain ; K12856 pre-mRNA-processin... 194 1e-49
sce:YHR165C PRP8, DBF3, DNA39, RNA8, SLT21, USA2; Component of... 185 4e-47
pfa:PFD0265w pre-mRNA splicing factor, putative; K12856 pre-mR... 142 5e-34
dre:570913 ugt5g1, zgc:175099; UDP glucuronosyltransferase 5 f... 30.0 3.0
ath:AT5G10610 CYP81K1; electron carrier/ heme binding / iron i... 29.3 4.2
mmu:69601 Dab2ip, 2310011D08Rik, AI480459, Aip1, KIAA1743, MGC... 29.3 4.8
hsa:153090 DAB2IP, AF9Q34, AIP1, DIP1/2, FLJ39072, KIAA1743; D... 29.3 5.0
bbo:BBOV_IV001040 21.m02803; hypothetical protein 28.9 5.7
dre:767756 ugt5a2, MGC153634, MGC153649, wu:fi04d07, zgc:15363... 28.9 6.0
dre:559348 syne2b, fc12c11, id:ibd5137, si:dkey-249p9.3, wu:fc... 28.5 7.3
> tgo:TGME49_031970 pre-mRNA splicing factor PRP8, putative ;
K12856 pre-mRNA-processing factor 8
Length=2538
Score = 268 bits (685), Expect = 4e-72, Method: Compositional matrix adjust.
Identities = 127/147 (86%), Positives = 136/147 (92%), Gaps = 0/147 (0%)
Query 1 WEISNRLPRSLTTLNWSSSFASVYSLDNPNLLFSMGGFECRILPKVRMQTEEFSQREGAW 60
WEI NRLPRS++TL WS+SFASVYS DNPNLLF+M GFE RILPK+R TEEFSQREG W
Sbjct 1357 WEIENRLPRSVSTLEWSNSFASVYSKDNPNLLFAMCGFEVRILPKIRTYTEEFSQREGVW 1416
Query 61 KLQNETTKELAAQAFLKVGDEGMRRFENRVRMVLMSSGATTFTKIANKWNTTLISLMTYF 120
KLQNE TKE+AAQAFLKVGDEGM+ FENRVR +LM+SGATTFTKIANKWNTTLISLMTYF
Sbjct 1417 KLQNEVTKEMAAQAFLKVGDEGMKHFENRVRQILMASGATTFTKIANKWNTTLISLMTYF 1476
Query 121 REAVIHTEALLDLLVKCENKIQTRIKI 147
REAVIHTEALLDLLVKCENKIQTRIKI
Sbjct 1477 REAVIHTEALLDLLVKCENKIQTRIKI 1503
> dre:393951 prpf8, MGC162871, MGC56504, id:ibd1257, ik:tdsubc_2a9,
im:7141966, tdsubc_2a9, wu:fb37c02, wu:fb73e06, xx:tdsubc_2a9,
zgc:56504; pre-mRNA processing factor 8; K12856 pre-mRNA-processing
factor 8
Length=2342
Score = 232 bits (591), Expect = 4e-61, Method: Compositional matrix adjust.
Identities = 106/147 (72%), Positives = 122/147 (82%), Gaps = 0/147 (0%)
Query 1 WEISNRLPRSLTTLNWSSSFASVYSLDNPNLLFSMGGFECRILPKVRMQTEEFSQREGAW 60
W+I NRLPRS+TT+ W +SF SVYS DNPNLLF+M GFECRILPK R EEF+ ++G W
Sbjct 1162 WDIKNRLPRSVTTIQWENSFVSVYSKDNPNLLFNMCGFECRILPKCRTSYEEFTHKDGVW 1221
Query 61 KLQNETTKELAAQAFLKVGDEGMRRFENRVRMVLMSSGATTFTKIANKWNTTLISLMTYF 120
LQNE TKE AQ FL+V DE M+RF NRVR +LM+SG+TTFTKI NKWNT LI LMTYF
Sbjct 1222 NLQNEVTKERTAQCFLRVDDESMQRFHNRVRQILMASGSTTFTKIVNKWNTALIGLMTYF 1281
Query 121 REAVIHTEALLDLLVKCENKIQTRIKI 147
REAV++T+ LLDLLVKCENKIQTRIKI
Sbjct 1282 REAVVNTQELLDLLVKCENKIQTRIKI 1308
> xla:379945 prpf8, MGC52804, hprp8, prp-8, prp8, prpc8, rp13;
PRP8 pre-mRNA processing factor 8 homolog; K12856 pre-mRNA-processing
factor 8
Length=2335
Score = 231 bits (590), Expect = 4e-61, Method: Compositional matrix adjust.
Identities = 106/147 (72%), Positives = 122/147 (82%), Gaps = 0/147 (0%)
Query 1 WEISNRLPRSLTTLNWSSSFASVYSLDNPNLLFSMGGFECRILPKVRMQTEEFSQREGAW 60
W+I NRLPRS+TT+ W +SF SVYS DNPNLLF+M GFECRILPK R EEF+ ++G W
Sbjct 1155 WDIKNRLPRSVTTVQWENSFVSVYSKDNPNLLFNMCGFECRILPKCRTSYEEFTHKDGVW 1214
Query 61 KLQNETTKELAAQAFLKVGDEGMRRFENRVRMVLMSSGATTFTKIANKWNTTLISLMTYF 120
LQNE TKE AQ FL+V DE M+RF NRVR +LM+SG+TTFTKI NKWNT LI LMTYF
Sbjct 1215 NLQNEVTKERTAQCFLRVDDESMQRFHNRVRQILMASGSTTFTKIVNKWNTALIGLMTYF 1274
Query 121 REAVIHTEALLDLLVKCENKIQTRIKI 147
REAV++T+ LLDLLVKCENKIQTRIKI
Sbjct 1275 REAVVNTQELLDLLVKCENKIQTRIKI 1301
> mmu:192159 Prpf8, AU019467, D11Bwg0410e, DBF3/PRP8, Prp8, Sfprp8l;
pre-mRNA processing factor 8; K12856 pre-mRNA-processing
factor 8
Length=2335
Score = 231 bits (590), Expect = 5e-61, Method: Compositional matrix adjust.
Identities = 106/147 (72%), Positives = 122/147 (82%), Gaps = 0/147 (0%)
Query 1 WEISNRLPRSLTTLNWSSSFASVYSLDNPNLLFSMGGFECRILPKVRMQTEEFSQREGAW 60
W+I NRLPRS+TT+ W +SF SVYS DNPNLLF+M GFECRILPK R EEF+ ++G W
Sbjct 1155 WDIKNRLPRSVTTVQWENSFVSVYSKDNPNLLFNMCGFECRILPKCRTSYEEFTHKDGVW 1214
Query 61 KLQNETTKELAAQAFLKVGDEGMRRFENRVRMVLMSSGATTFTKIANKWNTTLISLMTYF 120
LQNE TKE AQ FL+V DE M+RF NRVR +LM+SG+TTFTKI NKWNT LI LMTYF
Sbjct 1215 NLQNEVTKERTAQCFLRVDDESMQRFHNRVRQILMASGSTTFTKIVNKWNTALIGLMTYF 1274
Query 121 REAVIHTEALLDLLVKCENKIQTRIKI 147
REAV++T+ LLDLLVKCENKIQTRIKI
Sbjct 1275 REAVVNTQELLDLLVKCENKIQTRIKI 1301
> hsa:10594 PRPF8, HPRP8, PRP8, PRPC8, RP13; PRP8 pre-mRNA processing
factor 8 homolog (S. cerevisiae); K12856 pre-mRNA-processing
factor 8
Length=2335
Score = 231 bits (588), Expect = 9e-61, Method: Composition-based stats.
Identities = 106/147 (72%), Positives = 122/147 (82%), Gaps = 0/147 (0%)
Query 1 WEISNRLPRSLTTLNWSSSFASVYSLDNPNLLFSMGGFECRILPKVRMQTEEFSQREGAW 60
W+I NRLPRS+TT+ W +SF SVYS DNPNLLF+M GFECRILPK R EEF+ ++G W
Sbjct 1155 WDIKNRLPRSVTTVQWENSFVSVYSKDNPNLLFNMCGFECRILPKCRTSYEEFTHKDGVW 1214
Query 61 KLQNETTKELAAQAFLKVGDEGMRRFENRVRMVLMSSGATTFTKIANKWNTTLISLMTYF 120
LQNE TKE AQ FL+V DE M+RF NRVR +LM+SG+TTFTKI NKWNT LI LMTYF
Sbjct 1215 NLQNEVTKERTAQCFLRVDDESMQRFHNRVRQILMASGSTTFTKIVNKWNTALIGLMTYF 1274
Query 121 REAVIHTEALLDLLVKCENKIQTRIKI 147
REAV++T+ LLDLLVKCENKIQTRIKI
Sbjct 1275 REAVVNTQELLDLLVKCENKIQTRIKI 1301
> cel:C50C3.6 prp-8; yeast PRP (splicing factor) related family
member (prp-8); K12856 pre-mRNA-processing factor 8
Length=2329
Score = 228 bits (581), Expect = 5e-60, Method: Compositional matrix adjust.
Identities = 104/147 (70%), Positives = 119/147 (80%), Gaps = 0/147 (0%)
Query 1 WEISNRLPRSLTTLNWSSSFASVYSLDNPNLLFSMGGFECRILPKVRMQTEEFSQREGAW 60
W+I NRLPRS+TT+ W +SF SVYS DNPN+LF M GFECRILPK R EEF R+G W
Sbjct 1147 WDIKNRLPRSITTVEWENSFVSVYSKDNPNMLFDMSGFECRILPKCRTANEEFVHRDGVW 1206
Query 61 KLQNETTKELAAQAFLKVGDEGMRRFENRVRMVLMSSGATTFTKIANKWNTTLISLMTYF 120
LQNE TKE AQ FLKV +E + +F NR+R +LMSSG+TTFTKI NKWNT LI LMTYF
Sbjct 1207 NLQNEVTKERTAQCFLKVDEESLSKFHNRIRQILMSSGSTTFTKIVNKWNTALIGLMTYF 1266
Query 121 REAVIHTEALLDLLVKCENKIQTRIKI 147
REAV++T+ LLDLLVKCENKIQTRIKI
Sbjct 1267 REAVVNTQELLDLLVKCENKIQTRIKI 1293
> ath:AT1G80070 SUS2; SUS2 (ABNORMAL SUSPENSOR 2); K12856 pre-mRNA-processing
factor 8
Length=2382
Score = 224 bits (571), Expect = 7e-59, Method: Compositional matrix adjust.
Identities = 107/148 (72%), Positives = 120/148 (81%), Gaps = 1/148 (0%)
Query 1 WEISNRLPRSLTTLNWSSSFASVYSLDNPNLLFSMGGFECRILPKVRMQTEEFSQ-REGA 59
W++ NRLPRS+TTL W + F SVYS DNPNLLFSM GFE RILPK+RM E FS ++G
Sbjct 1201 WDMKNRLPRSITTLEWENGFVSVYSKDNPNLLFSMCGFEVRILPKIRMTQEAFSNTKDGV 1260
Query 60 WKLQNETTKELAAQAFLKVGDEGMRRFENRVRMVLMSSGATTFTKIANKWNTTLISLMTY 119
W LQNE TKE A AFL+V DE M+ FENRVR +LMSSG+TTFTKI NKWNT LI LMTY
Sbjct 1261 WNLQNEQTKERTAVAFLRVDDEHMKVFENRVRQILMSSGSTTFTKIVNKWNTALIGLMTY 1320
Query 120 FREAVIHTEALLDLLVKCENKIQTRIKI 147
FREA +HT+ LLDLLVKCENKIQTRIKI
Sbjct 1321 FREATVHTQELLDLLVKCENKIQTRIKI 1348
> ath:AT4G38780 splicing factor, putative
Length=2332
Score = 222 bits (566), Expect = 3e-58, Method: Compositional matrix adjust.
Identities = 105/148 (70%), Positives = 119/148 (80%), Gaps = 1/148 (0%)
Query 1 WEISNRLPRSLTTLNWSSSFASVYSLDNPNLLFSMGGFECRILPKVRMQTEEFSQ-REGA 59
W++ NRLPRS+TTL W + F SVYS DNPNLLFSM GFE R+LPK+RM E FS R+G
Sbjct 1153 WDMKNRLPRSITTLEWENGFVSVYSKDNPNLLFSMCGFEVRVLPKIRMGQEAFSSTRDGV 1212
Query 60 WKLQNETTKELAAQAFLKVGDEGMRRFENRVRMVLMSSGATTFTKIANKWNTTLISLMTY 119
W LQNE TKE A AFL+ DE M+ FENRVR +LMSSG+TTFTKI NKWNT LI LMTY
Sbjct 1213 WNLQNEQTKERTAVAFLRADDEHMKVFENRVRQILMSSGSTTFTKIVNKWNTALIGLMTY 1272
Query 120 FREAVIHTEALLDLLVKCENKIQTRIKI 147
FREA +HT+ LLDLLVKCENKIQTR+KI
Sbjct 1273 FREATVHTQELLDLLVKCENKIQTRVKI 1300
> bbo:BBOV_IV007790 23.m06497; processing splicing factor 8; K12856
pre-mRNA-processing factor 8
Length=2343
Score = 216 bits (551), Expect = 1e-56, Method: Compositional matrix adjust.
Identities = 103/147 (70%), Positives = 117/147 (79%), Gaps = 0/147 (0%)
Query 1 WEISNRLPRSLTTLNWSSSFASVYSLDNPNLLFSMGGFECRILPKVRMQTEEFSQREGAW 60
WE+ RLPRS+TTL W+ SF SVY DNPNLLF+M GFE RI PK+R +Q E W
Sbjct 1177 WEMQARLPRSVTTLEWNDSFVSVYGKDNPNLLFNMYGFEVRIFPKIRWLKSGVTQAEACW 1236
Query 61 KLQNETTKELAAQAFLKVGDEGMRRFENRVRMVLMSSGATTFTKIANKWNTTLISLMTYF 120
KLQNE TKEL+A A+L+V EGM FENRVR +LM+SG+TTFTKIANKWNT LI +MTY+
Sbjct 1237 KLQNERTKELSATAYLRVDAEGMSTFENRVRQILMASGSTTFTKIANKWNTALIGMMTYY 1296
Query 121 REAVIHTEALLDLLVKCENKIQTRIKI 147
REAVIHT LLDLLVKCENKIQTRIKI
Sbjct 1297 REAVIHTNELLDLLVKCENKIQTRIKI 1323
> tpv:TP03_0292 splicing factor Prp8; K12856 pre-mRNA-processing
factor 8
Length=2736
Score = 196 bits (499), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 101/156 (64%), Positives = 117/156 (75%), Gaps = 9/156 (5%)
Query 1 WEISNRLPRSLTTLNWSSSFASVYSLDNPNLLFSMGGFECRI--------LPKVRMQT-E 51
WE+ +RLPRS+TTL WS SF SVYS DNPNLLFS+ GFE RI + + T +
Sbjct 1574 WEMQSRLPRSITTLEWSDSFVSVYSKDNPNLLFSLCGFEVRIRRYGAANSVSDTTVGTVD 1633
Query 52 EFSQREGAWKLQNETTKELAAQAFLKVGDEGMRRFENRVRMVLMSSGATTFTKIANKWNT 111
E +W+LQN TK+L+A A+L+V +E M FENRVR +LMSSG+TTFTKIANKWNT
Sbjct 1634 TVKLSESSWRLQNMKTKQLSAIAYLRVSNESMSMFENRVRQILMSSGSTTFTKIANKWNT 1693
Query 112 TLISLMTYFREAVIHTEALLDLLVKCENKIQTRIKI 147
LISLMTYFREA IHT LLDLLVKCENKIQTRIKI
Sbjct 1694 ALISLMTYFREATIHTNELLDLLVKCENKIQTRIKI 1729
> cpv:cgd3_2890 Prp8. JAB/PAD domain ; K12856 pre-mRNA-processing
factor 8
Length=2379
Score = 194 bits (492), Expect = 1e-49, Method: Compositional matrix adjust.
Identities = 95/162 (58%), Positives = 114/162 (70%), Gaps = 15/162 (9%)
Query 1 WEISNRLPRSLTTLNWSSSFASVYSLDNPNLLFSMGGFECRILPKVRM------------ 48
WE+SNRLP+S+TTL W SF SVYS NPNLLFS+ GF RILP R+
Sbjct 1146 WELSNRLPKSITTLEWERSFVSVYSKSNPNLLFSLAGFSVRILPTCRIGKRTFEASNTSF 1205
Query 49 ---QTEEFSQREGAWKLQNETTKELAAQAFLKVGDEGMRRFENRVRMVLMSSGATTFTKI 105
+ ++ RE W+L N TKE+ + FL V + +R FENRVR +L++SG+ TFTKI
Sbjct 1206 IGNEDSQYYTRESTWQLSNNLTKEITSYVFLMVDESEIRNFENRVRQILITSGSATFTKI 1265
Query 106 ANKWNTTLISLMTYFREAVIHTEALLDLLVKCENKIQTRIKI 147
ANKWNT LI LMTYFREAVI+TE LLDLLV+CENKIQTRIKI
Sbjct 1266 ANKWNTCLIGLMTYFREAVIYTEKLLDLLVRCENKIQTRIKI 1307
> sce:YHR165C PRP8, DBF3, DNA39, RNA8, SLT21, USA2; Component
of the U4/U6-U5 snRNP complex, involved in the second catalytic
step of splicing; mutations of human Prp8 cause retinitis
pigmentosa; K12856 pre-mRNA-processing factor 8
Length=2413
Score = 185 bits (470), Expect = 4e-47, Method: Compositional matrix adjust.
Identities = 86/147 (58%), Positives = 117/147 (79%), Gaps = 1/147 (0%)
Query 1 WEISNRLPRSLTTLNWSSSFASVYSLDNPNLLFSMGGFECRILPKVRMQTEEFSQREGAW 60
WEI +R+P SLT++ W ++F SVYS +NPNLLFSM GFE RILP+ RM+ E S EG W
Sbjct 1228 WEIQSRVPTSLTSIKWENAFVSVYSKNNPNLLFSMCGFEVRILPRQRME-EVVSNDEGVW 1286
Query 61 KLQNETTKELAAQAFLKVGDEGMRRFENRVRMVLMSSGATTFTKIANKWNTTLISLMTYF 120
L +E TK+ A+A+LKV +E +++F++R+R +LM+SG+TTFTK+A KWNT+LISL TYF
Sbjct 1287 DLVDERTKQRTAKAYLKVSEEEIKKFDSRIRGILMASGSTTFTKVAAKWNTSLISLFTYF 1346
Query 121 REAVIHTEALLDLLVKCENKIQTRIKI 147
REA++ TE LLD+LVK E +IQ R+K+
Sbjct 1347 REAIVATEPLLDILVKGETRIQNRVKL 1373
> pfa:PFD0265w pre-mRNA splicing factor, putative; K12856 pre-mRNA-processing
factor 8
Length=3136
Score = 142 bits (357), Expect = 5e-34, Method: Composition-based stats.
Identities = 75/94 (79%), Positives = 83/94 (88%), Gaps = 0/94 (0%)
Query 54 SQREGAWKLQNETTKELAAQAFLKVGDEGMRRFENRVRMVLMSSGATTFTKIANKWNTTL 113
S +EG WKLQNE TKE+ A+A+LKV D M+RFENRVR +LMSSG+TTFTKIANKWNTTL
Sbjct 1863 SVKEGTWKLQNEMTKEITAEAYLKVSDNSMKRFENRVRQILMSSGSTTFTKIANKWNTTL 1922
Query 114 ISLMTYFREAVIHTEALLDLLVKCENKIQTRIKI 147
I LMTYFREAV+ TE LLDLLVKCENKIQTRIKI
Sbjct 1923 IGLMTYFREAVLDTEELLDLLVKCENKIQTRIKI 1956
Score = 77.0 bits (188), Expect = 2e-14, Method: Composition-based stats.
Identities = 34/49 (69%), Positives = 41/49 (83%), Gaps = 2/49 (4%)
Query 1 WEISNRLPRSLTTLNWS--SSFASVYSLDNPNLLFSMGGFECRILPKVR 47
WEI NR+PRSLT+L+W ++F SVYS DNPNLLFS+ GFE RILPK+R
Sbjct 1702 WEIQNRIPRSLTSLDWDHYNTFVSVYSKDNPNLLFSIAGFEVRILPKIR 1750
> dre:570913 ugt5g1, zgc:175099; UDP glucuronosyltransferase 5
family, polypeptide G1; K00699 glucuronosyltransferase [EC:2.4.1.17]
Length=528
Score = 30.0 bits (66), Expect = 3.0, Method: Composition-based stats.
Identities = 20/69 (28%), Positives = 31/69 (44%), Gaps = 12/69 (17%)
Query 29 PNLLFSMGGFECRILPKVRMQTEEFSQREGAWKL-----------QNETTKELAAQAFLK 77
PN+++ MGGF+C+ + + EEF Q G + T E A AF K
Sbjct 273 PNIIY-MGGFQCKPAQALPVDLEEFMQSSGEHGVVFMSLGAMVGALPRTITEAIASAFAK 331
Query 78 VGDEGMRRF 86
+ + M R+
Sbjct 332 IPQKVMWRY 340
> ath:AT5G10610 CYP81K1; electron carrier/ heme binding / iron
ion binding / monooxygenase/ oxygen binding
Length=500
Score = 29.3 bits (64), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 19/81 (23%), Positives = 39/81 (48%), Gaps = 3/81 (3%)
Query 37 GFECRILPKVRMQTEEFSQREGAWKLQNETTKELAAQAFLKVGDEGMRRFENRVR---MV 93
G E R++ RM+ E + +++N + + FLK+ + + + V +V
Sbjct 235 GLEKRVIDMQRMRDEYLQRLIDDIRMKNIDSSGSVVEKFLKLQESEPEFYADDVIKGIIV 294
Query 94 LMSSGATTFTKIANKWNTTLI 114
LM +G T + +A +W +L+
Sbjct 295 LMFNGGTDTSPVAMEWAVSLL 315
> mmu:69601 Dab2ip, 2310011D08Rik, AI480459, Aip1, KIAA1743, MGC144147,
mKIAA1743; disabled homolog 2 (Drosophila) interacting
protein
Length=1124
Score = 29.3 bits (64), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 17/43 (39%), Positives = 28/43 (65%), Gaps = 2/43 (4%)
Query 45 KVRMQTEEFSQREGAWKLQNETTKELA--AQAFLKVGDEGMRR 85
K+R+ T++ + E +K Q ETT++L QA L+ G+E +RR
Sbjct 983 KLRISTKKLEEYETLFKCQEETTQKLVLEYQARLEEGEERLRR 1025
> hsa:153090 DAB2IP, AF9Q34, AIP1, DIP1/2, FLJ39072, KIAA1743;
DAB2 interacting protein
Length=1132
Score = 29.3 bits (64), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 17/43 (39%), Positives = 28/43 (65%), Gaps = 2/43 (4%)
Query 45 KVRMQTEEFSQREGAWKLQNETTKELA--AQAFLKVGDEGMRR 85
K+R+ T++ + E +K Q ETT++L QA L+ G+E +RR
Sbjct 1020 KLRISTKKLEEYETLFKCQEETTQKLVLEYQARLEEGEERLRR 1062
> bbo:BBOV_IV001040 21.m02803; hypothetical protein
Length=178
Score = 28.9 bits (63), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 22/82 (26%), Positives = 35/82 (42%), Gaps = 7/82 (8%)
Query 1 WEISNRLPRSLTTLNWSSSFASVYSLDNPNLLFSMGGF--ECRILPKVRMQTEEFSQREG 58
W ++NR+ L S F SVY + L GG IL K++ +
Sbjct 85 WRVNNRVVSYLGACKLSEKFQSVY-----DDLHKNGGITVPSDILNKIKRSPSGPLSIDI 139
Query 59 AWKLQNETTKELAAQAFLKVGD 80
W L N+ K+L+++ +K D
Sbjct 140 LWSLSNKIAKQLSSKYGVKAND 161
> dre:767756 ugt5a2, MGC153634, MGC153649, wu:fi04d07, zgc:153634,
zgc:153649; UDP glucuronosyltransferase 5 family, polypeptide
A2
Length=524
Score = 28.9 bits (63), Expect = 6.0, Method: Composition-based stats.
Identities = 24/86 (27%), Positives = 38/86 (44%), Gaps = 4/86 (4%)
Query 29 PNLLFSMGGFECRILPKVRMQTEEFSQREGAWKLQNETTKELAAQAFLKVGDEGMRRFEN 88
PN+++ MGGF+C+ + EEF Q G + + + Q ++ DE F
Sbjct 269 PNVVY-MGGFQCKPAKPLPGDLEEFVQSSGEHGVITMSLGTVFGQLLSELNDEIAAAFAQ 327
Query 89 RVRMVLMSSGATTFTKIANKWNTTLI 114
+ V+ T + AN N TLI
Sbjct 328 LPQKVIWR---YTGPRPANLGNNTLI 350
> dre:559348 syne2b, fc12c11, id:ibd5137, si:dkey-249p9.3, wu:fc12c11,
wu:fc31e06, wu:fc62a06; spectrin repeat containing,
nuclear envelope 2b
Length=2742
Score = 28.5 bits (62), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 33/136 (24%), Positives = 56/136 (41%), Gaps = 18/136 (13%)
Query 10 SLTTLNWSSSFASVYSLDNPNLLFS--MGGFECR---ILPKVRMQTEEFSQREGAWKLQN 64
SL+ L+ + S Y L + LL + C+ + KV M+ +E + R AW + N
Sbjct 1699 SLSELSTMKTDLSQYILTDDVLLLQEQVEHLHCQWEELCLKVSMRKQEIADRLNAWIIFN 1758
Query 65 ETTKELAAQAFLKVGDEGMRRFENRVRMVLMSSGATTFTKIANKWNTTLISLMTYFREAV 124
E +EL E + + EN+V S + ++ K + + F E
Sbjct 1759 EKNRELC---------EWLTQMENKV----AHSAELSIEEMVEKLKKDCMEEINLFSENK 1805
Query 125 IHTEALLDLLVKCENK 140
H + L + L+ NK
Sbjct 1806 THLKQLGEQLITASNK 1821
Lambda K H
0.320 0.131 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2938175820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40