bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1586_orf2
Length=154
Score E
Sequences producing significant alignments: (Bits) Value
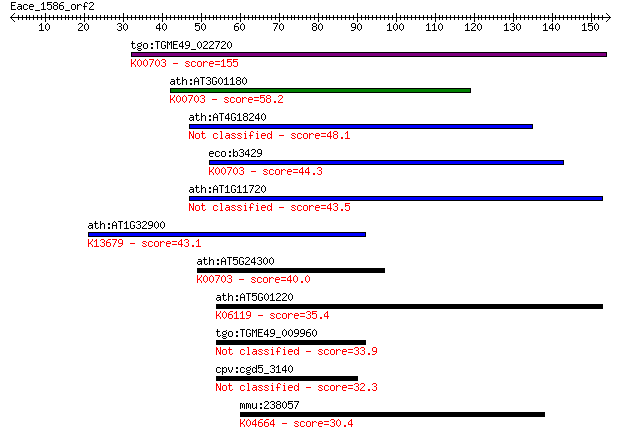
tgo:TGME49_022720 glycogen synthase, putative (EC:2.4.1.21); K... 155 3e-38
ath:AT3G01180 AtSS2; AtSS2 (starch synthase 2); transferase, t... 58.2 1e-08
ath:AT4G18240 ATSS4; ATSS4; transferase, transferring glycosyl... 48.1 1e-05
eco:b3429 glgA, ECK3415, JW3392; glycogen synthase (EC:2.4.1.2... 44.3 1e-04
ath:AT1G11720 ATSS3; ATSS3 (starch synthase 3); starch synthas... 43.5 3e-04
ath:AT1G32900 starch synthase, putative; K13679 granule-bound ... 43.1 4e-04
ath:AT5G24300 SSI1; SSI1 (SUPPRESSOR OF SALICYLIC ACID INSENSI... 40.0 0.003
ath:AT5G01220 SQD2; SQD2 (sulfoquinovosyldiacylglycerol 2); UD... 35.4 0.080
tgo:TGME49_009960 glycan synthetase, putative (EC:2.4.1.21 2.4... 33.9 0.20
cpv:cgd5_3140 LPS glycosyltransferase of possible cyanobacteri... 32.3 0.69
mmu:238057 Gdf7, BMP12; growth differentiation factor 7; K0466... 30.4 2.3
> tgo:TGME49_022720 glycogen synthase, putative (EC:2.4.1.21);
K00703 starch synthase [EC:2.4.1.21]
Length=1350
Score = 155 bits (393), Expect = 3e-38, Method: Compositional matrix adjust.
Identities = 83/134 (61%), Positives = 92/134 (68%), Gaps = 12/134 (8%)
Query 32 PAEFLGRPNEFFTKGAHVNYGADFGLMPSAFEPGGIVQHEFFISGTPVIAFHTGGLKDTV 91
P F P EFF GA VN GADFGLMPS FEPGGIVQHEFFI+GTPV+AF TGGLKDTV
Sbjct 1044 PHSFWADPGEFFCNGALVNLGADFGLMPSLFEPGGIVQHEFFIAGTPVVAFRTGGLKDTV 1103
Query 92 VE--FVPAAGTG----------NGFSFMNYTSGDLLYAMERAIRVFDDKEKYTLLRQLAR 139
E P G G NGF+F YT+GD L+A+ERA+RVF D+ KY LR AR
Sbjct 1104 REGSVTPGLGGGKISVESARQNNGFTFDAYTAGDFLFAIERALRVFSDRAKYEQLRANAR 1163
Query 140 QSVVSCECSSWAWL 153
SVVSCE S+ AWL
Sbjct 1164 ASVVSCEESARAWL 1177
> ath:AT3G01180 AtSS2; AtSS2 (starch synthase 2); transferase,
transferring glycosyl groups; K00703 starch synthase [EC:2.4.1.21]
Length=792
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 33/78 (42%), Positives = 46/78 (58%), Gaps = 1/78 (1%)
Query 42 FFTKGAH-VNYGADFGLMPSAFEPGGIVQHEFFISGTPVIAFHTGGLKDTVVEFVPAAGT 100
F K AH + GAD LMPS FEP G+ Q GT + GGL+DTV +F P + T
Sbjct 669 FSVKTAHRITAGADILLMPSRFEPCGLNQLYAMNYGTIPVVHAVGGLRDTVQQFDPYSET 728
Query 101 GNGFSFMNYTSGDLLYAM 118
G G++F + +G L++A+
Sbjct 729 GLGWTFDSAEAGKLIHAL 746
> ath:AT4G18240 ATSS4; ATSS4; transferase, transferring glycosyl
groups (EC:2.4.1.21)
Length=1040
Score = 48.1 bits (113), Expect = 1e-05, Method: Composition-based stats.
Identities = 34/94 (36%), Positives = 47/94 (50%), Gaps = 7/94 (7%)
Query 47 AHVNYGA-DFGLMPSAFEPGGIVQHEFFISGTPVIAFHTGGLKDTVVEF----VPAAGTG 101
+H Y A D ++PS FEP G+ Q G+ IA TGGL D+V + +P
Sbjct 912 SHTIYAASDLFIIPSIFEPCGLTQMIAMRYGSIPIARKTGGLNDSVFDIDDDTIPTQFQ- 970
Query 102 NGFSFMNYTSGDLLYAMERAIRVF-DDKEKYTLL 134
NGF+F YA+ERA + D+EK+ L
Sbjct 971 NGFTFQTADEQGFNYALERAFNHYKKDEEKWMRL 1004
> eco:b3429 glgA, ECK3415, JW3392; glycogen synthase (EC:2.4.1.21);
K00703 starch synthase [EC:2.4.1.21]
Length=477
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 34/94 (36%), Positives = 49/94 (52%), Gaps = 6/94 (6%)
Query 52 GADFGLMPSAFEPGGIVQHEFFISGTPVIAFHTGGLKDTVVEFV---PAAGTGNGFSFMN 108
GAD L+PS FEP G+ Q GT + TGGL DTV + A G +GF F +
Sbjct 366 GADVILVPSRFEPCGLTQLYGLKYGTLPLVRRTGGLADTVSDCSLENLADGVASGFVFED 425
Query 109 YTSGDLLYAMERAIRVFDDKEKYTLLRQLARQSV 142
+ LL A+ RA ++ + +L R + RQ++
Sbjct 426 SNAWSLLRAIRRAFVLW---SRPSLWRFVQRQAM 456
> ath:AT1G11720 ATSS3; ATSS3 (starch synthase 3); starch synthase/
transferase, transferring glycosyl groups (EC:2.4.1.21)
Length=1025
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 37/116 (31%), Positives = 53/116 (45%), Gaps = 18/116 (15%)
Query 47 AHVNY-GADFGLMPSAFEPGGIVQHEFFISGTPVIAFHTGGLKDTVVEFVPAAGTG---- 101
+H+ Y GADF L+PS FEP G+ Q G + TGGL DTV +
Sbjct 902 SHLIYAGADFILVPSIFEPCGLTQLIAMRYGAVPVVRKTGGLFDTVFDVDHDKERAQAQV 961
Query 102 ---NGFSFMNYTSGDLLYAMERAIRVFDDKEKY--TLLRQLARQSVVSCECSSWAW 152
NGFSF + + YA+ RAI + D ++ +L + + Q W+W
Sbjct 962 LEPNGFSFDGADAPGVDYALNRAISAWYDGREWFNSLCKTVMEQ--------DWSW 1009
> ath:AT1G32900 starch synthase, putative; K13679 granule-bound
starch synthase [EC:2.4.1.242]
Length=610
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 28/75 (37%), Positives = 40/75 (53%), Gaps = 4/75 (5%)
Query 21 RYKDDGAAVEIPAEFLGRP---NEFFTKGAH-VNYGADFGLMPSAFEPGGIVQHEFFISG 76
+ K + +E+ +F G+ +F AH + GADF ++PS FEP G++Q G
Sbjct 440 KKKMEAQILELEEKFPGKAVGVAKFNVPLAHMITAGADFIIVPSRFEPCGLIQLHAMRYG 499
Query 77 TPVIAFHTGGLKDTV 91
T I TGGL DTV
Sbjct 500 TVPIVASTGGLVDTV 514
> ath:AT5G24300 SSI1; SSI1 (SUPPRESSOR OF SALICYLIC ACID INSENSITIVITY
1); starch synthase/ transferase, transferring glycosyl
groups; K00703 starch synthase [EC:2.4.1.21]
Length=652
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 22/48 (45%), Positives = 26/48 (54%), Gaps = 0/48 (0%)
Query 49 VNYGADFGLMPSAFEPGGIVQHEFFISGTPVIAFHTGGLKDTVVEFVP 96
+ G D LMPS FEP G+ Q GT + TGGL+DTV F P
Sbjct 529 ITAGCDILLMPSRFEPCGLNQLYAMRYGTIPVVHGTGGLRDTVENFNP 576
> ath:AT5G01220 SQD2; SQD2 (sulfoquinovosyldiacylglycerol 2);
UDP-glycosyltransferase/ UDP-sulfoquinovose:DAG sulfoquinovosyltransferase/
transferase, transferring glycosyl groups; K06119
sulfoquinovosyltransferase [EC:2.4.1.-]
Length=510
Score = 35.4 bits (80), Expect = 0.080, Method: Compositional matrix adjust.
Identities = 29/100 (29%), Positives = 44/100 (44%), Gaps = 13/100 (13%)
Query 54 DFGLMPSAFEPGGIVQHEFFISGTPVIAFHTGGLKDTVVEFVPAAGTGN-GFSFMNYTSG 112
D +MPS E G+V E SG PV+A GG+ D +P G GF F G
Sbjct 378 DVFVMPSESETLGLVVLEAMSSGLPVVAARAGGIPD----IIPEDQEGKTGFLF---NPG 430
Query 113 DLLYAMERAIRVFDDKEKYTLLRQLARQSVVSCECSSWAW 152
D+ + + + D+E ++ + AR+ E + W
Sbjct 431 DVEDCVTKLRTLLHDRETREIIGKAARE-----ETEKYDW 465
> tgo:TGME49_009960 glycan synthetase, putative (EC:2.4.1.21 2.4.1.18)
Length=1707
Score = 33.9 bits (76), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 18/38 (47%), Positives = 25/38 (65%), Gaps = 0/38 (0%)
Query 54 DFGLMPSAFEPGGIVQHEFFISGTPVIAFHTGGLKDTV 91
D ++PS EP GIV E + SG PV+A ++GG +D V
Sbjct 1343 DAVVVPSRNEPFGIVVLEAWSSGKPVVATNSGGPRDFV 1380
> cpv:cgd5_3140 LPS glycosyltransferase of possible cyanobacterial
origin
Length=2069
Score = 32.3 bits (72), Expect = 0.69, Method: Composition-based stats.
Identities = 16/36 (44%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 54 DFGLMPSAFEPGGIVQHEFFISGTPVIAFHTGGLKD 89
D ++PS EP GIV E + +G PV+A +GG +D
Sbjct 1537 DAVVVPSRNEPFGIVVLEGWTAGKPVVATTSGGPRD 1572
> mmu:238057 Gdf7, BMP12; growth differentiation factor 7; K04664
growth differentiation factor 5/6/7
Length=453
Score = 30.4 bits (67), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 25/91 (27%), Positives = 35/91 (38%), Gaps = 13/91 (14%)
Query 60 SAFEPGGIVQHEFFIS-------GTPVIAFHTGGLKDTVVEFVPAAGTGNGFSFMNYTSG 112
S F G +V H F +S PV A G DT+ F A G F F +
Sbjct 75 SGFRNGSVVPHHFMMSLYRSLAGRAPVAAASGHGRVDTITGFTDQATQGQSFLFDVSSLS 134
Query 113 DLLYAMERAIRVF------DDKEKYTLLRQL 137
+ + +RV D++ TLL +L
Sbjct 135 EADEVVNAELRVLRRRSPEPDRDSATLLPRL 165
Lambda K H
0.323 0.139 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3321543300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40