bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1430_orf2
Length=176
Score E
Sequences producing significant alignments: (Bits) Value
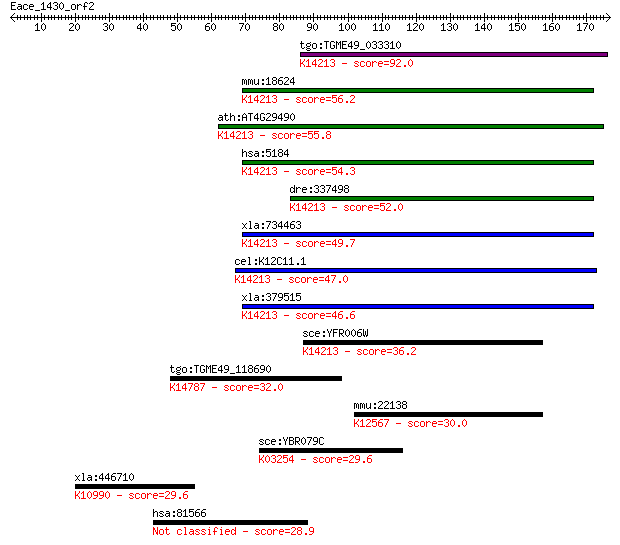
tgo:TGME49_033310 prolidase, putative (EC:3.4.13.9); K14213 Xa... 92.0 1e-18
mmu:18624 Pepd, Pep-4, Pep4; peptidase D (EC:3.4.13.9); K14213... 56.2 6e-08
ath:AT4G29490 aminopeptidase/ manganese ion binding; K14213 Xa... 55.8 7e-08
hsa:5184 PEPD, MGC10905, PROLIDASE; peptidase D (EC:3.4.13.9);... 54.3 2e-07
dre:337498 pepd, cb1000, fj78g11, wu:fj78g11; peptidase D (EC:... 52.0 1e-06
xla:734463 pepd, MGC115123; peptidase D (EC:3.4.13.9); K14213 ... 49.7 5e-06
cel:K12C11.1 hypothetical protein; K14213 Xaa-Pro dipeptidase ... 47.0 3e-05
xla:379515 hypothetical protein MGC64570; K14213 Xaa-Pro dipep... 46.6 5e-05
sce:YFR006W Putative X-Pro aminopeptidase; green fluorescent p... 36.2 0.052
tgo:TGME49_118690 RNA recognition motif-containing protein ; K... 32.0 1.2
mmu:22138 Ttn, 1100001C23Rik, 2310036G12Rik, 2310057K23Rik, 23... 30.0 4.4
sce:YBR079C RPG1, TIF32; Rpg1p; K03254 translation initiation ... 29.6 5.3
xla:446710 rmi1, MGC83955; RMI1, RecQ mediated genome instabil... 29.6 5.7
hsa:81566 CSRNP2, C12orf2, C12orf22, FAM130A1, FLJ25576, TAIP-... 28.9 9.0
> tgo:TGME49_033310 prolidase, putative (EC:3.4.13.9); K14213
Xaa-Pro dipeptidase [EC:3.4.13.9]
Length=525
Score = 92.0 bits (227), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 39/90 (43%), Positives = 56/90 (62%), Gaps = 0/90 (0%)
Query 86 VAAFLEGGTTDEWMLYSSDCNKAAFRQEAFFQHLIGINEPDIYAAYILASEELVLFTPKI 145
VAAF +GG+ +W YS+DC+KA FRQE FF++L G+NE D++ + + VLF P
Sbjct 129 VAAFFQGGSAADWAFYSADCDKAVFRQEQFFRYLFGVNEADVFGLLDFSRRQAVLFVPWT 188
Query 146 PEDALRFMGPPKDNNFYVSRYAVTDVVEVK 175
+ RFMGPP+ +Y+ RY + V K
Sbjct 189 SPEYQRFMGPPRAAEWYMQRYGLDGAVVYK 218
> mmu:18624 Pepd, Pep-4, Pep4; peptidase D (EC:3.4.13.9); K14213
Xaa-Pro dipeptidase [EC:3.4.13.9]
Length=493
Score = 56.2 bits (134), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 31/103 (30%), Positives = 49/103 (47%), Gaps = 3/103 (2%)
Query 69 RRNFIEAVRAKGLAKEGVAAFLEGGTTDEWMLYSSDCNKAAFRQEAFFQHLIGINEPDIY 128
R+ E +R G + A L+GG +E Y +D FRQE+FF G+ E Y
Sbjct 27 RQRLCERLRKNGAVQAASAVVLQGG--EEMQRYCTD-TSIIFRQESFFHWAFGVVESGCY 83
Query 129 AAYILASEELVLFTPKIPEDALRFMGPPKDNNFYVSRYAVTDV 171
+ + + LF P++P+ +MG ++ +YAV DV
Sbjct 84 GVIDVDTGKSTLFVPRLPDSYATWMGKIHSKEYFKEKYAVDDV 126
> ath:AT4G29490 aminopeptidase/ manganese ion binding; K14213
Xaa-Pro dipeptidase [EC:3.4.13.9]
Length=486
Score = 55.8 bits (133), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 37/113 (32%), Positives = 60/113 (53%), Gaps = 8/113 (7%)
Query 62 KEKLEEVRRNFIEAVRAKGLAKEGVAAFLEGGTTDEWMLYSSDCNKAAFRQEAFFQHLIG 121
K+ LE +RR + R+ +G L+GG +E Y +D + FRQE++F +L G
Sbjct 19 KKLLESIRRQLSSSNRSL----DGFV-LLQGG--EEKNRYCTD-HTELFRQESYFAYLFG 70
Query 122 INEPDIYAAYILASEELVLFTPKIPEDALRFMGPPKDNNFYVSRYAVTDVVEV 174
+ EPD Y A + S + +LF P++P+D ++G K + + Y V V V
Sbjct 71 VREPDFYGAIDIGSGKSILFIPRLPDDYAVWLGEIKPLSHFKETYMVDMVFYV 123
> hsa:5184 PEPD, MGC10905, PROLIDASE; peptidase D (EC:3.4.13.9);
K14213 Xaa-Pro dipeptidase [EC:3.4.13.9]
Length=493
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 31/103 (30%), Positives = 47/103 (45%), Gaps = 3/103 (2%)
Query 69 RRNFIEAVRAKGLAKEGVAAFLEGGTTDEWMLYSSDCNKAAFRQEAFFQHLIGINEPDIY 128
R+ E +R + G L+GG +E Y +D FRQE+FF G+ EP Y
Sbjct 27 RQRLCERLRKNPAVQAGSIVVLQGG--EETQRYCTD-TGVLFRQESFFHWAFGVTEPGCY 83
Query 129 AAYILASEELVLFTPKIPEDALRFMGPPKDNNFYVSRYAVTDV 171
+ + + LF P++P +MG + +YAV DV
Sbjct 84 GVIDVDTGKSTLFVPRLPASHATWMGKIHSKEHFKEKYAVDDV 126
> dre:337498 pepd, cb1000, fj78g11, wu:fj78g11; peptidase D (EC:3.4.13.9);
K14213 Xaa-Pro dipeptidase [EC:3.4.13.9]
Length=496
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/89 (33%), Positives = 48/89 (53%), Gaps = 3/89 (3%)
Query 83 KEGVAAFLEGGTTDEWMLYSSDCNKAAFRQEAFFQHLIGINEPDIYAAYILASEELVLFT 142
+ G L+GG ++ Y +D ++ FRQE+FF G+ E D Y A + S++ +LF
Sbjct 42 QAGSVVVLQGG--EQKQRYCTDTDET-FRQESFFHWSFGVTEADCYGAIDVDSKKSLLFV 98
Query 143 PKIPEDALRFMGPPKDNNFYVSRYAVTDV 171
PK+PE +MG + +YAV +V
Sbjct 99 PKLPESYATWMGEIFPPGHFKEKYAVDEV 127
> xla:734463 pepd, MGC115123; peptidase D (EC:3.4.13.9); K14213
Xaa-Pro dipeptidase [EC:3.4.13.9]
Length=498
Score = 49.7 bits (117), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 28/103 (27%), Positives = 48/103 (46%), Gaps = 3/103 (2%)
Query 69 RRNFIEAVRAKGLAKEGVAAFLEGGTTDEWMLYSSDCNKAAFRQEAFFQHLIGINEPDIY 128
R+ + +R +G L+GG + Y +D FRQE+FF G+ EP Y
Sbjct 27 RKRLCDILRQNKDLPKGSIVLLQGGEATQR--YCTD-TGTLFRQESFFHWTFGVTEPGCY 83
Query 129 AAYILASEELVLFTPKIPEDALRFMGPPKDNNFYVSRYAVTDV 171
A + + + + F PK+PE +MG + +YA+ ++
Sbjct 84 GAVDVDTGKSIAFIPKLPESYAVWMGKIHPPEHFKEKYAIDEI 126
> cel:K12C11.1 hypothetical protein; K14213 Xaa-Pro dipeptidase
[EC:3.4.13.9]
Length=498
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 32/107 (29%), Positives = 54/107 (50%), Gaps = 5/107 (4%)
Query 67 EVRRNFIEAVRAKGLAKEGVAAFLEGGTTDEWMLYSSDCNKAAFRQEAFFQHLIGINEPD 126
E R ++A+++K A V L+GG E Y++D FRQE++F G+NE +
Sbjct 19 ENRHRLVDALKSKVPANSVV--LLQGGV--EKNRYNTDAADLPFRQESYFFWTFGVNESE 74
Query 127 IYAAY-ILASEELVLFTPKIPEDALRFMGPPKDNNFYVSRYAVTDVV 172
Y A + + + LF P++ + G + F+ +YAV +VV
Sbjct 75 FYGAIDVRSGGKTTLFAPRLDPSYAIWDGKINNEQFFKEKYAVDEVV 121
> xla:379515 hypothetical protein MGC64570; K14213 Xaa-Pro dipeptidase
[EC:3.4.13.9]
Length=498
Score = 46.6 bits (109), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 28/103 (27%), Positives = 48/103 (46%), Gaps = 3/103 (2%)
Query 69 RRNFIEAVRAKGLAKEGVAAFLEGGTTDEWMLYSSDCNKAAFRQEAFFQHLIGINEPDIY 128
R+ + ++ G L+GG +E Y +D FRQE+FF G+ E Y
Sbjct 27 RKRLCDRLKQNKDVPTGSFVLLQGG--EETQRYCTD-TGILFRQESFFHWTFGVIEAGCY 83
Query 129 AAYILASEELVLFTPKIPEDALRFMGPPKDNNFYVSRYAVTDV 171
A + + + +LF PK+PE +MG + +YA+ ++
Sbjct 84 GAVDVNTGKSILFIPKLPESYAVWMGKIHPPEHFKEKYAIDEI 126
> sce:YFR006W Putative X-Pro aminopeptidase; green fluorescent
protein (GFP)-fusion protein localizes to the cytoplasm; YFR006W
is not an essential gene (EC:3.4.-.-); K14213 Xaa-Pro
dipeptidase [EC:3.4.13.9]
Length=535
Score = 36.2 bits (82), Expect = 0.052, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 35/70 (50%), Gaps = 2/70 (2%)
Query 87 AAFLEGGTTDEWMLYSSDCNKAAFRQEAFFQHLIGINEPDIYAAYILASEELVLFTPKIP 146
AF G E Y D NK FRQ +F HL G++ P + ++++L LF P I
Sbjct 97 TAFFIAGEELEGNKYC-DTNKD-FRQNRYFYHLSGVDIPASAILFNCSTDKLTLFLPNID 154
Query 147 EDALRFMGPP 156
E+ + + G P
Sbjct 155 EEDVIWSGMP 164
> tgo:TGME49_118690 RNA recognition motif-containing protein ;
K14787 multiple RNA-binding domain-containing protein 1
Length=997
Score = 32.0 bits (71), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 31/61 (50%), Gaps = 14/61 (22%)
Query 48 HLLLREYLTSPLI-----------HKEKLEEVRRNFIEAVRAKGLAKEGVAAFLEGGTTD 96
HL R+Y PL +E++E R+ +E VRA +A+ GVA +GG+ +
Sbjct 638 HLAYRQYKNVPLFLEKAPVNVFVEREEEIETARKGAVEEVRAAKIAERGVA---DGGSKN 694
Query 97 E 97
E
Sbjct 695 E 695
> mmu:22138 Ttn, 1100001C23Rik, 2310036G12Rik, 2310057K23Rik,
2310074I15Rik, AF006999, AV006427, D330041I19Rik, D830007G01Rik,
L56, mdm, shru; titin (EC:2.7.11.1); K12567 titin [EC:2.7.11.1]
Length=33467
Score = 30.0 bits (66), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 18/55 (32%), Positives = 25/55 (45%), Gaps = 7/55 (12%)
Query 102 SSDCNKAAFRQEAFFQHLIGINEPDIYAAYILASEELVLFTPKIPEDALRFMGPP 156
SS CNK +FR E + E IY ++A E + P DA++ PP
Sbjct 24209 SSKCNKTSFRVE-------NLTEGAIYYFRVMAENEFGVGVPTETSDAVKASEPP 24256
> sce:YBR079C RPG1, TIF32; Rpg1p; K03254 translation initiation
factor 3 subunit A
Length=964
Score = 29.6 bits (65), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 19/45 (42%), Positives = 23/45 (51%), Gaps = 3/45 (6%)
Query 74 EAVRAKGLAKEGVAAFLEGGTTDEWML---YSSDCNKAAFRQEAF 115
E RA L K+ + LEGG T E +L Y SD + A F EA
Sbjct 107 EQTRADELQKQEIDDDLEGGVTPENLLISVYESDQSVAGFNDEAI 151
> xla:446710 rmi1, MGC83955; RMI1, RecQ mediated genome instability
1, homolog; K10990 protein RMI1 homolog
Length=557
Score = 29.6 bits (65), Expect = 5.7, Method: Composition-based stats.
Identities = 16/37 (43%), Positives = 21/37 (56%), Gaps = 2/37 (5%)
Query 20 QLLETTQQADRPWLAWPTTWKGLQESD--DHLLLREY 54
Q+ TTQQ+ +PW A PT LQ +D H+ EY
Sbjct 115 QVTCTTQQSQKPWEAKPTRMLMLQLTDGTQHIQAMEY 151
> hsa:81566 CSRNP2, C12orf2, C12orf22, FAM130A1, FLJ25576, TAIP-12;
cysteine-serine-rich nuclear protein 2
Length=543
Score = 28.9 bits (63), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 43 QESDDHLLLREYLTSPLIHKEKLEEVRRNFIEAVRAKGLAKEGVA 87
QE + +LRE+L +H +K++ + +E+V A GL + V+
Sbjct 114 QEVNHREILREHLKEEKLHAKKMKLTKNGTVESVEADGLTLDDVS 158
Lambda K H
0.318 0.135 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4600750868
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40