bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1428_orf1
Length=110
Score E
Sequences producing significant alignments: (Bits) Value
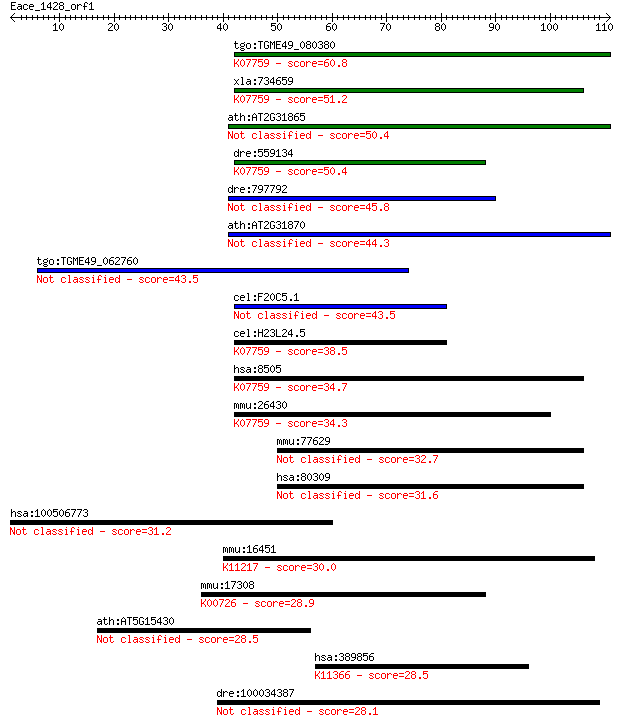
tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143); K0... 60.8 1e-09
xla:734659 parg, MGC115697; poly (ADP-ribose) glycohydrolase; ... 51.2 9e-07
ath:AT2G31865 poly (ADP-ribose) glycohydrolase (PARG) family p... 50.4 1e-06
dre:559134 poly(ADP-ribose) glycohydrolase 63 kDa-like; K07759... 50.4 2e-06
dre:797792 si:dkey-259k14.2 45.8 3e-05
ath:AT2G31870 TEJ; TEJ (Sanskrit for 'bright'); poly(ADP-ribos... 44.3 9e-05
tgo:TGME49_062760 poly(ADP-ribose) glycohydrolase, putative 43.5 2e-04
cel:F20C5.1 pme-3; Poly(ADP-ribose) Metabolism Enzyme family m... 43.5 2e-04
cel:H23L24.5 pme-4; Poly(ADP-ribose) Metabolism Enzyme family ... 38.5 0.005
hsa:8505 PARG, FLJ54459, FLJ60257, FLJ60456, PARG99; poly (ADP... 34.7 0.078
mmu:26430 Parg, AI413217; poly (ADP-ribose) glycohydrolase (EC... 34.3 0.11
mmu:77629 Sphkap, 4930544G21Rik, A930009L15Rik, AI852220, mKIA... 32.7 0.28
hsa:80309 SPHKAP, DKFZp781H143, DKFZp781J171, KIAA1678, MGC132... 31.6 0.73
hsa:100506773 glycine N-acyltransferase-like protein 1-like 31.2 0.90
mmu:16451 Jak1, AA960307, C130039L05Rik, MGC37919; Janus kinas... 30.0 1.8
mmu:17308 Mgat1, Mgat-1; mannoside acetylglucosaminyltransfera... 28.9 4.6
ath:AT5G15430 calmodulin-binding protein-related 28.5 5.3
hsa:389856 USP27X, USP22L, USP27; ubiquitin specific peptidase... 28.5 5.3
dre:100034387 sphkap, si:dkey-223n17.6, si:dkey-99o15.1; SPHK1... 28.1 6.3
> tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143);
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=553
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 45/69 (65%), Gaps = 0/69 (0%)
Query 42 WGCVIFGGDNELKTLMQWISAAAAGRELHYKAFGDTRLSHMQVTLHAIREKFPTTRDLFE 101
WGC +F GD +LKTL+QW++A+ AGR + + F + + + + + +R+ + T +L++
Sbjct 469 WGCGVFNGDAQLKTLIQWLAASYAGRSMKFYTFSNKSVDGLGLVISKLRQSYATVGNLYK 528
Query 102 AIVQSVQQR 110
AI ++ +R
Sbjct 529 AIQNALSRR 537
> xla:734659 parg, MGC115697; poly (ADP-ribose) glycohydrolase;
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=759
Score = 51.2 bits (121), Expect = 9e-07, Method: Composition-based stats.
Identities = 25/65 (38%), Positives = 37/65 (56%), Gaps = 1/65 (1%)
Query 42 WGCVIFGGDNELKTLMQWISAAAAGRELHYKAFGDTRL-SHMQVTLHAIREKFPTTRDLF 100
WGC FGGD LK L+Q ++AA GR+L Y FGD L + + + EK T D++
Sbjct 667 WGCGAFGGDPRLKALIQLLAAAEVGRDLVYFTFGDRELMKDIYLMYSFLTEKNKTVGDIY 726
Query 101 EAIVQ 105
+++
Sbjct 727 SMLIE 731
> ath:AT2G31865 poly (ADP-ribose) glycohydrolase (PARG) family
protein
Length=522
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 41/71 (57%), Gaps = 1/71 (1%)
Query 41 KWGCVIFGGDNELKTLMQWISAAAAGRE-LHYKAFGDTRLSHMQVTLHAIREKFPTTRDL 99
WGC +FGGD ELK ++QW++ + +GR + Y FG L ++ + + + T DL
Sbjct 433 NWGCGVFGGDPELKIMLQWLAISQSGRPFMSYYTFGLQALQNLNQVIEMVALQEMTVGDL 492
Query 100 FEAIVQSVQQR 110
++ +V+ +R
Sbjct 493 WKKLVEYSSER 503
> dre:559134 poly(ADP-ribose) glycohydrolase 63 kDa-like; K07759
poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=777
Score = 50.4 bits (119), Expect = 2e-06, Method: Composition-based stats.
Identities = 23/46 (50%), Positives = 28/46 (60%), Gaps = 0/46 (0%)
Query 42 WGCVIFGGDNELKTLMQWISAAAAGRELHYKAFGDTRLSHMQVTLH 87
WGC FGGD LK L+Q ++AA AGR++ Y FGD L LH
Sbjct 664 WGCGAFGGDTRLKALLQLMAAAEAGRDVAYFTFGDEALMRDVQDLH 709
> dre:797792 si:dkey-259k14.2
Length=609
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 20/49 (40%), Positives = 30/49 (61%), Gaps = 0/49 (0%)
Query 41 KWGCVIFGGDNELKTLMQWISAAAAGRELHYKAFGDTRLSHMQVTLHAI 89
WGC F GD +LK L+Q ++AA R++ Y FG+T L++ +H I
Sbjct 515 NWGCGAFRGDPKLKALLQLMAAAVVDRDVAYFTFGNTHLANELQKMHDI 563
> ath:AT2G31870 TEJ; TEJ (Sanskrit for 'bright'); poly(ADP-ribose)
glycohydrolase
Length=547
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 27/72 (37%), Positives = 41/72 (56%), Gaps = 3/72 (4%)
Query 41 KWGCVIFGGDNELKTLMQWISAAAAGRE-LHYKAFGDTRLSHM-QVTLHAIREKFPTTRD 98
WGC +FGGD ELK +QW++A+ R + Y FG L ++ QVT + K+ T D
Sbjct 450 NWGCGVFGGDPELKATIQWLAASQTRRPFISYYTFGVEALRNLDQVTKWILSHKW-TVGD 508
Query 99 LFEAIVQSVQQR 110
L+ +++ QR
Sbjct 509 LWNMMLEYSAQR 520
> tgo:TGME49_062760 poly(ADP-ribose) glycohydrolase, putative
Length=952
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 30/77 (38%), Positives = 41/77 (53%), Gaps = 12/77 (15%)
Query 6 SLSTLRGPLRRLQLALRRSRLLSS--GLQGNPTRGEFK-------WGCVIFGGDNELKTL 56
S+ST G LR+ + R R +S L P +G K WGC +F GD +LK L
Sbjct 791 SVSTPDGALRQNE---ERDRFSASEATLAPGPHQGLVKRPFATGNWGCGVFKGDPQLKFL 847
Query 57 MQWISAAAAGRELHYKA 73
+QW++A+ GR L Y A
Sbjct 848 LQWLAASLVGRRLIYHA 864
> cel:F20C5.1 pme-3; Poly(ADP-ribose) Metabolism Enzyme family
member (pme-3)
Length=764
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 18/39 (46%), Positives = 25/39 (64%), Gaps = 0/39 (0%)
Query 42 WGCVIFGGDNELKTLMQWISAAAAGRELHYKAFGDTRLS 80
WGC F GD LK ++Q I+A A R LH+ +FG+ L+
Sbjct 662 WGCGAFNGDKPLKFIIQVIAAGVADRPLHFCSFGEPELA 700
> cel:H23L24.5 pme-4; Poly(ADP-ribose) Metabolism Enzyme family
member (pme-4); K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=485
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 17/39 (43%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 42 WGCVIFGGDNELKTLMQWISAAAAGRELHYKAFGDTRLS 80
WGC F G+ LK L+Q I+ + R L + FGDT L+
Sbjct 396 WGCGAFRGNKPLKFLIQVIACGISDRPLQFCTFGDTELA 434
> hsa:8505 PARG, FLJ54459, FLJ60257, FLJ60456, PARG99; poly (ADP-ribose)
glycohydrolase (EC:3.2.1.143); K07759 poly(ADP-ribose)
glycohydrolase [EC:3.2.1.143]
Length=976
Score = 34.7 bits (78), Expect = 0.078, Method: Composition-based stats.
Identities = 25/65 (38%), Positives = 41/65 (63%), Gaps = 1/65 (1%)
Query 42 WGCVIFGGDNELKTLMQWISAAAAGRELHYKAFGDTRLSHMQVTLHA-IREKFPTTRDLF 100
WGC FGGD LK L+Q ++AAAA R++ Y FGD+ L ++H + E+ T D++
Sbjct 870 WGCGAFGGDARLKALIQILAAAAAERDVVYFTFGDSELMRDIYSMHIFLTERKLTVGDVY 929
Query 101 EAIVQ 105
+ +++
Sbjct 930 KLLLR 934
> mmu:26430 Parg, AI413217; poly (ADP-ribose) glycohydrolase (EC:3.2.1.143);
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=961
Score = 34.3 bits (77), Expect = 0.11, Method: Composition-based stats.
Identities = 26/58 (44%), Positives = 34/58 (58%), Gaps = 4/58 (6%)
Query 42 WGCVIFGGDNELKTLMQWISAAAAGRELHYKAFGDTRLSHMQVTLHAIREKFPTTRDL 99
WGC FGGD LK L+Q ++AAAA R++ Y FGD+ L ++H F T R L
Sbjct 863 WGCGAFGGDARLKALIQILAAAAAERDVVYFTFGDSELMRDIYSMHT----FLTERKL 916
> mmu:77629 Sphkap, 4930544G21Rik, A930009L15Rik, AI852220, mKIAA1678;
SPHK1 interactor, AKAP domain containing
Length=1658
Score = 32.7 bits (73), Expect = 0.28, Method: Composition-based stats.
Identities = 16/57 (28%), Positives = 32/57 (56%), Gaps = 1/57 (1%)
Query 50 DNELKTLMQWISAAAAG-RELHYKAFGDTRLSHMQVTLHAIREKFPTTRDLFEAIVQ 105
D EL+ +QWI+A+ G +++K ++R+ + +++K D+F A+VQ
Sbjct 1579 DAELRATLQWIAASELGIPTIYFKKSQESRIEKFLDVVKLVQQKSWKVGDIFHAVVQ 1635
> hsa:80309 SPHKAP, DKFZp781H143, DKFZp781J171, KIAA1678, MGC132614,
MGC132616, SKIP; SPHK1 interactor, AKAP domain containing
Length=1700
Score = 31.6 bits (70), Expect = 0.73, Method: Composition-based stats.
Identities = 16/57 (28%), Positives = 29/57 (50%), Gaps = 1/57 (1%)
Query 50 DNELKTLMQWISAAAAG-RELHYKAFGDTRLSHMQVTLHAIREKFPTTRDLFEAIVQ 105
D EL+ +QWI+A+ G +++K + R+ + + K D+F A+VQ
Sbjct 1621 DAELRATLQWIAASELGIPTIYFKKSQENRIEKFLDVVQLVHRKSWKVGDIFHAVVQ 1677
> hsa:100506773 glycine N-acyltransferase-like protein 1-like
Length=548
Score = 31.2 bits (69), Expect = 0.90, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 29/63 (46%), Gaps = 4/63 (6%)
Query 1 CPYWQS--LSTLRGPLRRLQLAL-RRSRLLSSGLQGNPT-RGEFKWGCVIFGGDNELKTL 56
CP WQS + + P L R S L S+G QG P R E C+++ G EL
Sbjct 471 CPSWQSKVWESAKAPRASHSAQLPRSSSLASTGRQGTPMRRKEQGSDCLVYPGTRELSPW 530
Query 57 MQW 59
+W
Sbjct 531 TEW 533
> mmu:16451 Jak1, AA960307, C130039L05Rik, MGC37919; Janus kinase
1 (EC:2.7.10.2); K11217 Janus kinase 1 [EC:2.7.10.2]
Length=1153
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 17/68 (25%), Positives = 31/68 (45%), Gaps = 3/68 (4%)
Query 40 FKWGCVIFGGDNELKTLMQWISAAAAGRELHYKAFGDTRLSHMQVTLHAIREKFPTTRDL 99
+W C F DN L T+ + + G + +K F + + +LH + FP+ RDL
Sbjct 465 LRWSCTDF--DNILMTVTCFEKSEVLGGQKQFKNF-QIEVQKGRYSLHGSMDHFPSLRDL 521
Query 100 FEAIVQSV 107
+ + +
Sbjct 522 MNHLKKQI 529
> mmu:17308 Mgat1, Mgat-1; mannoside acetylglucosaminyltransferase
1 (EC:2.4.1.101); K00726 alpha-1,3-mannosyl-glycoprotein
beta-1,2-N-acetylglucosaminyltransferase [EC:2.4.1.101]
Length=447
Score = 28.9 bits (63), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 23/52 (44%), Gaps = 1/52 (1%)
Query 36 TRGEFKWGCVIFGGDNELKTLMQWISAAAAGRELHYKAFGDTRLSHMQVTLH 87
T G WG +IF G N L L W + A GR A GD S + +H
Sbjct 6 TAGLVLWGAIIFVGWNALLLLFFW-TRPAPGRLPSDSALGDDPASLTREVIH 56
> ath:AT5G15430 calmodulin-binding protein-related
Length=478
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 25/40 (62%), Gaps = 1/40 (2%)
Query 17 LQLALRRSRLLSSGLQGN-PTRGEFKWGCVIFGGDNELKT 55
++L +RR +++ G +GN P + +FK G +I G D K+
Sbjct 355 MKLRIRRGKIIDFGSEGNSPRKLKFKRGKIISGADTTSKS 394
> hsa:389856 USP27X, USP22L, USP27; ubiquitin specific peptidase
27, X-linked (EC:3.4.19.12); K11366 ubiquitin carboxyl-terminal
hydrolase 22/27/51 [EC:3.1.2.15]
Length=438
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 57 MQWISAAAAGRELHYKAFGDTRLSHMQVTLHAIREKFPT 95
++ I+ G L +A T +SH Q ++ + EKFPT
Sbjct 11 IEQIAKEEQGEALKLQASTSTEVSHQQCSVPGLGEKFPT 49
> dre:100034387 sphkap, si:dkey-223n17.6, si:dkey-99o15.1; SPHK1
interactor, AKAP domain containing
Length=1596
Score = 28.1 bits (61), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 21/71 (29%), Positives = 33/71 (46%), Gaps = 5/71 (7%)
Query 39 EFKWGCVIFGGDNELKTLMQWISAAAAG-RELHYKAFGDTRLSHMQVTLHAIREKFPTTR 97
+ + CV D EL+ +QWISA+ G L+++ L+ + L +K
Sbjct 1506 DLEPDCV----DGELRAALQWISASELGVPALYFRKTHQHNLTKLHRVLQLAGQKAWRVG 1561
Query 98 DLFEAIVQSVQ 108
DLF A+ Q Q
Sbjct 1562 DLFSAVAQFCQ 1572
Lambda K H
0.324 0.136 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2067351240
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40