bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1415_orf1
Length=160
Score E
Sequences producing significant alignments: (Bits) Value
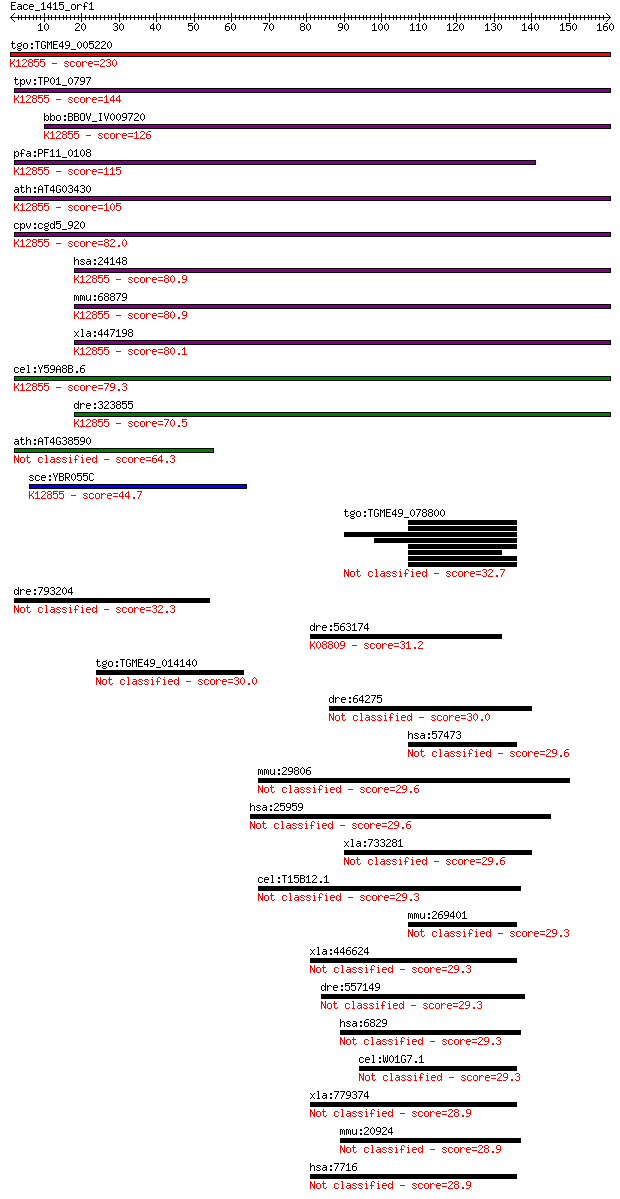
tgo:TGME49_005220 U5 snRNP-associated 102 kDa protein, putativ... 230 2e-60
tpv:TP01_0797 hypothetical protein; K12855 pre-mRNA-processing... 144 8e-35
bbo:BBOV_IV009720 23.m06014; u5 snRNP-associated subunit, puta... 126 4e-29
pfa:PF11_0108 U5 snRNP-associated protein, putative; K12855 pr... 115 7e-26
ath:AT4G03430 EMB2770 (EMBRYO DEFECTIVE 2770); RNA splicing fa... 105 5e-23
cpv:cgd5_920 Pre-mRNA splicing factor Pro1/Prp6. HAT repeat pr... 82.0 7e-16
hsa:24148 PRPF6, ANT-1, C20orf14, Prp6, TOM, U5-102K, hPrp6; P... 80.9 2e-15
mmu:68879 Prpf6, 1190003A07Rik, 2610031L17Rik, ANT-1, MGC11655... 80.9 2e-15
xla:447198 prpf6, MGC80263; PRP6 pre-mRNA processing factor 6 ... 80.1 3e-15
cel:Y59A8B.6 hypothetical protein; K12855 pre-mRNA-processing ... 79.3 4e-15
dre:323855 c20orf14, fc12b02, prpf6, wu:fa05f07, wu:fc12b02, z... 70.5 2e-12
ath:AT4G38590 BGAL14; glycosyl hydrolase family 35 protein 64.3 1e-10
sce:YBR055C PRP6, RNA6, TSM7269; Prp6p; K12855 pre-mRNA-proces... 44.7 1e-04
tgo:TGME49_078800 hypothetical protein 32.7 0.46
dre:793204 hypothetical LOC793204 32.3 0.75
dre:563174 si:ch211-195m20.1; K08809 striated muscle-specific ... 31.2 1.5
tgo:TGME49_014140 hypothetical protein 30.0 3.4
dre:64275 supt5h, Spt5, etID10129.15, etID37101.15, fb37g01, f... 30.0 3.7
hsa:57473 ZNF512B, GM632, KIAA1196, MGC149845, MGC149846; zinc... 29.6 4.1
mmu:29806 Limd1, AW822033, D9Ertd192e; LIM domains containing 1 29.6
hsa:25959 KANK2, ANKRD25, DKFZp434N161, FLJ20004, KIAA1518, MG... 29.6 4.5
xla:733281 supt5h, spt5, spt5h, tat-ct1; suppressor of Ty 5 ho... 29.6 4.9
cel:T15B12.1 hypothetical protein 29.3 5.1
mmu:269401 Znf512b, Gm632; zinc finger protein 512B 29.3 5.3
xla:446624 vezf1-a, MGC82370, db1, vezf1, vezf1a, znf161; vasc... 29.3 5.4
dre:557149 si:ch211-244b2.1 29.3 6.3
hsa:6829 SUPT5H, FLJ34157, SPT5, SPT5H, Tat-CT1; suppressor of... 29.3 6.4
cel:W01G7.1 daf-5; abnormal DAuer Formation family member (daf-5) 29.3 6.6
xla:779374 vezf1-b, MGC83349, db1, vezf1b, znf161; vascular en... 28.9 6.6
mmu:20924 Supt5h, AL033283, AL033321, AU019549, Spt5; suppress... 28.9 6.8
hsa:7716 VEZF1, DB1, ZNF161; vascular endothelial zinc finger 1 28.9
> tgo:TGME49_005220 U5 snRNP-associated 102 kDa protein, putative
; K12855 pre-mRNA-processing factor 6
Length=985
Score = 230 bits (586), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 118/164 (71%), Positives = 135/164 (82%), Gaps = 6/164 (3%)
Query 1 GRRRSRREARLKEEIAKLRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDYTLKRKQK 60
RR+SRRE RLKEEIAK+RAEKPTIHQQFADLK L +VT+EEWE+IP +GDYTLKRKQK
Sbjct 168 ARRKSRRENRLKEEIAKMRAEKPTIHQQFADLKRSLATVTKEEWEAIPSVGDYTLKRKQK 227
Query 61 KQQMLTPASDRGILEARNRESFSNSIASAGSATPIGFGMATPLMAGGLSTPLGLQTPLGI 120
K QM + A D +L+ RN S+SNSIASAGSATPIGFGM TPLM G++TPLGLQTPLG+
Sbjct 228 KPQMFSMAPDSLLLQGRNSTSYSNSIASAGSATPIGFGMQTPLM--GMATPLGLQTPLGL 285
Query 121 QTPL----GLRTPMGSSTPSLNDLGEARGTVLSVKLDKVMDSIS 160
+TPL G+ TPSLNDLGEARGTVLSVKLDKVMD++S
Sbjct 286 RTPLLGSGSGTGSSGAGTPSLNDLGEARGTVLSVKLDKVMDNLS 329
> tpv:TP01_0797 hypothetical protein; K12855 pre-mRNA-processing
factor 6
Length=1032
Score = 144 bits (364), Expect = 8e-35, Method: Compositional matrix adjust.
Identities = 84/160 (52%), Positives = 106/160 (66%), Gaps = 13/160 (8%)
Query 2 RRRSRREARLKEEIAKLRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDYTLKRK-QK 60
RR+SRRE LK EI+KLR+EKPTIH+Q A K L+++++EEWESIP IGDY+LKRK QK
Sbjct 123 RRKSRREQNLKNEISKLRSEKPTIHEQLAQYKRNLSTLSKEEWESIPYIGDYSLKRKQQK 182
Query 61 KQQMLTPASDRGILEARNRESFSNSIASAGSATPIGFGMATPLMAGGLSTPLGLQTPLGI 120
K Q P D I +R ++SI G+ TP+GF +TPL G TPLG+QTP G
Sbjct 183 KLQTYVPPPDSLIYSSRANMQHTSSI---GTETPLGF--STPLGIMGAKTPLGMQTPGGF 237
Query 121 QTPLGLRTPMGSSTPSLNDLGEARGTVLSVKLDKVMDSIS 160
T G ++ +LN LGEARG VLS LDKV D++S
Sbjct 238 TTTSGRKS-------TLNLLGEARGEVLSSTLDKVTDNLS 270
> bbo:BBOV_IV009720 23.m06014; u5 snRNP-associated subunit, putaitve;
K12855 pre-mRNA-processing factor 6
Length=1040
Score = 126 bits (316), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 76/157 (48%), Positives = 99/157 (63%), Gaps = 22/157 (14%)
Query 10 RLKEEIAKLRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDYTLKRKQ--KKQQMLTP 67
+++ E+ K RA+KPTIHQQ A LK L +++ EEWESIP IGDY+ KRKQ K+Q T
Sbjct 132 QIRTEVNKHRADKPTIHQQLAPLKRDLKNLSLEEWESIPSIGDYSFKRKQQNKRQHQYTA 191
Query 68 ASDRGILEARNRESFSNSIASAGSATPIGFGMATPLMAGGLSTPLGLQTPLGIQTPLGLR 127
A D + A+ +SI G++TP+GF STPLG+ G TP G+R
Sbjct 192 APDSLLYSAKVHMQSESSI---GTSTPLGF-----------STPLGIMG--GSATPSGVR 235
Query 128 TPMGSS----TPSLNDLGEARGTVLSVKLDKVMDSIS 160
+ + S+ T SLNDLGEARG VLS+ LDKVMD+IS
Sbjct 236 SSLISATSGDTSSLNDLGEARGAVLSITLDKVMDNIS 272
> pfa:PF11_0108 U5 snRNP-associated protein, putative; K12855
pre-mRNA-processing factor 6
Length=1329
Score = 115 bits (288), Expect = 7e-26, Method: Composition-based stats.
Identities = 75/146 (51%), Positives = 95/146 (65%), Gaps = 10/146 (6%)
Query 2 RRRSRREARLKEEIAKLRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDYTLKRKQKK 61
RR+SRRE +LKEEI+K+RA KPTI QQF DLK L +VT EEWESIP + Y+ K+KQKK
Sbjct 112 RRKSRRENKLKEEISKMRATKPTITQQFGDLKKNLANVTIEEWESIPTVLQYSSKQKQKK 171
Query 62 QQM-LTPASDRGILEARNRE----SFSNSIAS-AGSATPIGFGMATPLMAGGLSTPLGLQ 115
Q PA D I+ N +F+NS +S +G TPIG G + L G+ TPLGL+
Sbjct 172 VQKNYLPAPDSLIMSRINESNIHLNFNNSASSQSGHKTPIGLGYQSSL---GVQTPLGLR 228
Query 116 TPLGIQTPL-GLRTPMGSSTPSLNDL 140
TP G+Q L GL+TP+ SL+ L
Sbjct 229 TPYGLQNSLSGLKTPLSGLQNSLSGL 254
> ath:AT4G03430 EMB2770 (EMBRYO DEFECTIVE 2770); RNA splicing
factor, transesterification mechanism; K12855 pre-mRNA-processing
factor 6
Length=1029
Score = 105 bits (263), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 64/159 (40%), Positives = 88/159 (55%), Gaps = 29/159 (18%)
Query 2 RRRSRREARLKEEIAKLRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDYTLKRKQKK 61
RR+ RREA+LKEEI K RA P I +QFADLK L +++ +EW+SIP+IGDY+L+ K+KK
Sbjct 212 RRKDRREAKLKEEIEKYRASNPKITEQFADLKRKLHTLSADEWDSIPEIGDYSLRNKKKK 271
Query 62 QQMLTPASDRGILEARNRESFSNSIASAGSATPIGFGMATPLMAGGLSTPLGLQTPLGIQ 121
+ P D + +A+ + ++ A AGG TP G Q
Sbjct 272 FESFVPIPDTLLEKAKKEKELVMALDPKSRA------------AGGSETPWG-------Q 312
Query 122 TPLGLRTPMGSSTPSLNDLGEARGTVLSVKLDKVMDSIS 160
TP+ L +GE RGTVLS+KLD + DS+S
Sbjct 313 TPV----------TDLTAVGEGRGTVLSLKLDNLSDSVS 341
> cpv:cgd5_920 Pre-mRNA splicing factor Pro1/Prp6. HAT repeat
protein ; K12855 pre-mRNA-processing factor 6
Length=923
Score = 82.0 bits (201), Expect = 7e-16, Method: Composition-based stats.
Identities = 52/164 (31%), Positives = 80/164 (48%), Gaps = 47/164 (28%)
Query 2 RRRSRREARLKEEIAKLRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDYTLKRKQKK 61
RR+ ++E +++EEI K+R +PT+ +QF+ LK L V EEW+ IP+ GDY +K K+ K
Sbjct 105 RRKKQKEKKIREEILKVREHRPTLQEQFSGLKKSLGDVKIEEWDQIPEPGDYYIKNKKPK 164
Query 62 QQMLTP-----ASDRGILEARNRESFSNSIASAGSATPIGFGMATPLMAGGLSTPLGLQT 116
+ P +S + + E +++ SNS L+T L
Sbjct 165 LFLPVPDEIIQSSHKNLFETLTQKNCSNS---------------------ELNTEL---- 199
Query 117 PLGIQTPLGLRTPMGSSTPSLNDLGEARGTVLSVKLDKVMDSIS 160
T LN+LG A+G +LS+KLDK M S+S
Sbjct 200 -----------------TTELNELGTAKGNILSLKLDKAMGSVS 226
> hsa:24148 PRPF6, ANT-1, C20orf14, Prp6, TOM, U5-102K, hPrp6;
PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae);
K12855 pre-mRNA-processing factor 6
Length=941
Score = 80.9 bits (198), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 57/144 (39%), Positives = 84/144 (58%), Gaps = 12/144 (8%)
Query 18 LRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDYTLKRKQK-KQQMLTPASDRGILEA 76
R E+P I QQF+DLK L VT+EEW SIP++GD KR++ + + LTP D A
Sbjct 131 YRMERPKIQQQFSDLKRKLAEVTEEEWLSIPEVGDARNKRQRNPRYEKLTPVPDSFF--A 188
Query 77 RNRESFSNSIASAGSATPIGFGMATPLMAGGLSTPLGLQTPLGIQTPLGLRTPMGSSTPS 136
++ ++ N + T G G+ TP GGL+TP P G+ TP GL TP G+
Sbjct 189 KHLQTGENHTSVDPRQTQFG-GLNTP-YPGGLNTPY----PGGM-TP-GLMTP-GTGELD 239
Query 137 LNDLGEARGTVLSVKLDKVMDSIS 160
+ +G+AR T++ ++L +V DS+S
Sbjct 240 MRKIGQARNTLMDMRLSQVSDSVS 263
> mmu:68879 Prpf6, 1190003A07Rik, 2610031L17Rik, ANT-1, MGC11655,
MGC36967, MGC38351, U5-102K; PRP6 pre-mRNA splicing factor
6 homolog (yeast); K12855 pre-mRNA-processing factor 6
Length=941
Score = 80.9 bits (198), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 57/144 (39%), Positives = 84/144 (58%), Gaps = 12/144 (8%)
Query 18 LRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDYTLKRKQK-KQQMLTPASDRGILEA 76
R E+P I QQF+DLK L VT+EEW SIP++GD KR++ + + LTP D A
Sbjct 131 YRMERPKIQQQFSDLKRKLAEVTEEEWLSIPEVGDARNKRQRNPRYEKLTPVPDSFF--A 188
Query 77 RNRESFSNSIASAGSATPIGFGMATPLMAGGLSTPLGLQTPLGIQTPLGLRTPMGSSTPS 136
++ ++ N + T G G+ TP GGL+TP P G+ TP GL TP G+
Sbjct 189 KHLQTGENHTSVDPRQTQFG-GLNTP-YPGGLNTPY----PGGM-TP-GLMTP-GTGELD 239
Query 137 LNDLGEARGTVLSVKLDKVMDSIS 160
+ +G+AR T++ ++L +V DS+S
Sbjct 240 MRKIGQARNTLMDMRLSQVSDSVS 263
> xla:447198 prpf6, MGC80263; PRP6 pre-mRNA processing factor
6 homolog; K12855 pre-mRNA-processing factor 6
Length=948
Score = 80.1 bits (196), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 56/144 (38%), Positives = 83/144 (57%), Gaps = 12/144 (8%)
Query 18 LRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDYTLKRKQK-KQQMLTPASDRGILEA 76
R E+P I QQF+DLK L VT+EEW SIP++GD KR++ + + LTP D A
Sbjct 138 YRMERPKIQQQFSDLKRKLAEVTEEEWLSIPEVGDARNKRQRNPRHEKLTPVPDSFF--A 195
Query 77 RNRESFSNSIASAGSATPIGFGMATPLMAGGLSTPLGLQTPLGIQTPLGLRTPMGSSTPS 136
++ ++ N + G G+ TP GGL++P P G+ TP GL TP GS
Sbjct 196 KHLQTGENHTSVDPRQNQFG-GLNTPF-PGGLNSPY----PGGM-TP-GLMTP-GSGDLD 246
Query 137 LNDLGEARGTVLSVKLDKVMDSIS 160
+ +G+AR T++ ++L +V DS+S
Sbjct 247 MRKIGQARNTLMDMRLSQVSDSVS 270
> cel:Y59A8B.6 hypothetical protein; K12855 pre-mRNA-processing
factor 6
Length=968
Score = 79.3 bits (194), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 55/169 (32%), Positives = 83/169 (49%), Gaps = 17/169 (10%)
Query 2 RRRSRREARLKEEIAKLRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDYTLKRKQK- 60
R + RRE + KE + ++P I Q F DLK L VT++EW++IP++GD K K+
Sbjct 128 RHKERREKKYKELVELFHKDRPKIQQGFQDLKRQLAEVTEDEWQAIPEVGDMRNKAKRNA 187
Query 61 KQQMLTPASDRGILEARNRESFSNSIASAGSATPIGFGMATPLMAGGLSTPLGLQTPLGI 120
+ + TP D I N S+SI S G G+ TP +G +ST G
Sbjct 188 RAEKFTPVPDSIIAMNMNYGQMSHSIDS-------GNGLTTPFASGFMSTLGGGGGAAAA 240
Query 121 QTPLGLRTP-MGSSTPS--------LNDLGEARGTVLSVKLDKVMDSIS 160
G+ TP S PS L +G+AR ++ ++L +V DS++
Sbjct 241 GAKSGIMTPGWKSDIPSSGTSTDLDLVKIGQARNKIMDMQLTQVSDSVT 289
> dre:323855 c20orf14, fc12b02, prpf6, wu:fa05f07, wu:fc12b02,
zgc:65913; c20orf14 homolog (H. sapiens); K12855 pre-mRNA-processing
factor 6
Length=944
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 52/152 (34%), Positives = 77/152 (50%), Gaps = 33/152 (21%)
Query 18 LRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDYTLKRKQK-KQQMLTPASDRGILEA 76
R E+P I QQF+DLK L+ VT++EW SIP++GD KR++ + + LTP D
Sbjct 139 YRMERPKIQQQFSDLKRKLSEVTEDEWLSIPEVGDARNKRQRNPRYEKLTPVPD------ 192
Query 77 RNRESFSNSIASAGSATPIGFGMATPLMA--GGLSTPLGLQTPLGIQTPLGLRTPM-GSS 133
F+ + + + T + PL GGL+TP P L TP G
Sbjct 193 ---SFFAKHLQTGENHTSVD-----PLQGQFGGLNTPF----------PGVLNTPYPGGM 234
Query 134 TPSLNDL-----GEARGTVLSVKLDKVMDSIS 160
TP +L G+AR T++ ++L +V DS+S
Sbjct 235 TPDAGELDMRKIGQARNTLMDMRLSQVSDSVS 266
> ath:AT4G38590 BGAL14; glycosyl hydrolase family 35 protein
Length=1052
Score = 64.3 bits (155), Expect = 1e-10, Method: Composition-based stats.
Identities = 26/53 (49%), Positives = 39/53 (73%), Gaps = 0/53 (0%)
Query 2 RRRSRREARLKEEIAKLRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDYT 54
RR+ RREA+LK+EI RA P + QF DL L +++++EW+SIP+IG+Y+
Sbjct 996 RRKDRREAKLKQEIENYRASNPKVSGQFVDLTRKLHTLSEDEWDSIPEIGNYS 1048
> sce:YBR055C PRP6, RNA6, TSM7269; Prp6p; K12855 pre-mRNA-processing
factor 6
Length=899
Score = 44.7 bits (104), Expect = 1e-04, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 33/58 (56%), Gaps = 0/58 (0%)
Query 6 RREARLKEEIAKLRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDYTLKRKQKKQQ 63
+++ R E+ + + +QFADLK L +VT+ EW IPD D+T + K+ + Q
Sbjct 81 KKKKRANEKDDDNSVDSSNVKRQFADLKESLAAVTESEWMDIPDATDFTRRNKRNRIQ 138
> tgo:TGME49_078800 hypothetical protein
Length=1373
Score = 32.7 bits (73), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 14/29 (48%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 107 GLSTPLGLQTPLGIQTPLGLRTPMGSSTP 135
G+ TP G Q PLG+Q P G++ P G P
Sbjct 612 GVQTPPGEQAPLGVQAPPGVQAPPGVQAP 640
Score = 32.7 bits (73), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 14/29 (48%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 107 GLSTPLGLQTPLGIQTPLGLRTPMGSSTP 135
G+ TP G Q PLG+Q P G++ P G P
Sbjct 774 GVQTPPGEQAPLGVQAPPGVQAPPGVQAP 802
Score = 32.3 bits (72), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 20/50 (40%), Positives = 25/50 (50%), Gaps = 7/50 (14%)
Query 90 GSATPIGF----GMATPLMAGGLSTPLGLQTPLGIQTPLGLRTPMGSSTP 135
G TP G G+ TP G PLG+Q P G+Q P G++ P G P
Sbjct 600 GVQTPPGVQALPGVQTPP---GEQAPLGVQAPPGVQAPPGVQAPPGEQAP 646
Score = 31.6 bits (70), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 21/38 (55%), Gaps = 3/38 (7%)
Query 98 GMATPLMAGGLSTPLGLQTPLGIQTPLGLRTPMGSSTP 135
G+ TP G PLG+Q P G+Q P G++ P G P
Sbjct 774 GVQTPP---GEQAPLGVQAPPGVQAPPGVQAPPGEQAP 808
Score = 29.6 bits (65), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 107 GLSTPLGLQTPLGIQTPLGLRTPMGSSTP 135
G+ TP G+Q G+Q P G++TP G P
Sbjct 708 GVQTPPGVQALPGVQAPPGVQTPPGEQAP 736
Score = 29.3 bits (64), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 13/25 (52%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 107 GLSTPLGLQTPLGIQTPLGLRTPMG 131
G+ P G+QTP G Q P GL+ P G
Sbjct 720 GVQAPPGVQTPPGEQAPPGLQAPPG 744
Score = 28.5 bits (62), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 12/29 (41%), Positives = 16/29 (55%), Gaps = 0/29 (0%)
Query 107 GLSTPLGLQTPLGIQTPLGLRTPMGSSTP 135
G+ P G+Q P G+Q P G + P G P
Sbjct 624 GVQAPPGVQAPPGVQAPPGEQAPPGVEAP 652
Score = 28.5 bits (62), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 12/29 (41%), Positives = 16/29 (55%), Gaps = 0/29 (0%)
Query 107 GLSTPLGLQTPLGIQTPLGLRTPMGSSTP 135
G+ P G+Q P G+Q P G + P G P
Sbjct 786 GVQAPPGVQAPPGVQAPPGEQAPPGVEAP 814
> dre:793204 hypothetical LOC793204
Length=332
Score = 32.3 bits (72), Expect = 0.75, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 28/52 (53%), Gaps = 7/52 (13%)
Query 2 RRRSRREARLKEEIAKLRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDY 53
RRR R+ ARL + A+K T + +MG S+T E SIP +G+Y
Sbjct 271 RRRKRQAARLAH-VQSYFAQKAT------ETQMGCNSITDESSTSIPRLGEY 315
> dre:563174 si:ch211-195m20.1; K08809 striated muscle-specific
serine/threonine protein kinase [EC:2.7.11.1]
Length=3712
Score = 31.2 bits (69), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 25/51 (49%), Gaps = 4/51 (7%)
Query 81 SFSNSIASAGSATPIGFGMATPLMAGGLSTPLGLQTPLGIQTPLGLRTPMG 131
+ S + S +P+G G TP TP G TP G TP G RTP+G
Sbjct 3342 ALSPPVVLVTSLSPVGEGTGTP----NRMTPSGRATPSGRITPSGRRTPIG 3388
> tgo:TGME49_014140 hypothetical protein
Length=1975
Score = 30.0 bits (66), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 17/39 (43%), Positives = 24/39 (61%), Gaps = 1/39 (2%)
Query 24 TIHQQFADLKMGLTSVTQEEWESIPDIGDYTLKRKQKKQ 62
I QQ A LK+ L +TQ W+SI + D+ L++ KKQ
Sbjct 1050 AIRQQKAALKL-LKEITQWGWKSIYEHEDFFLEKLVKKQ 1087
> dre:64275 supt5h, Spt5, etID10129.15, etID37101.15, fb37g01,
fb95e05, fog, foggy, wu:fb37g01, wu:fb95e05, wu:fc10f02; suppressor
of Ty 5 homolog (S. cerevisiae)
Length=1084
Score = 30.0 bits (66), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 24/58 (41%), Positives = 29/58 (50%), Gaps = 5/58 (8%)
Query 86 IASAGSATPIGFGMATPLMAGGLSTPLGLQTPL--GIQTP--LGLRTPMGSSTPSLND 139
I GS TP+ +G TPL G + G QTPL G +TP G P +TPS D
Sbjct 775 IYGTGSRTPM-YGSQTPLHDGSRTPHYGSQTPLHDGSRTPGQSGAWDPNNPNTPSRPD 831
> hsa:57473 ZNF512B, GM632, KIAA1196, MGC149845, MGC149846; zinc
finger protein 512B
Length=892
Score = 29.6 bits (65), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 12/29 (41%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 107 GLSTPLGLQTPLGIQTPLGLRTPMGSSTP 135
G+S P+G+ P+ I P+G+ P+G S P
Sbjct 187 GVSKPIGVSKPVTIGKPVGVSKPIGISKP 215
> mmu:29806 Limd1, AW822033, D9Ertd192e; LIM domains containing
1
Length=668
Score = 29.6 bits (65), Expect = 4.3, Method: Composition-based stats.
Identities = 22/83 (26%), Positives = 34/83 (40%), Gaps = 3/83 (3%)
Query 67 PASDRGILEARNRESFSNSIASAGSATPIGFGMATPLMAGGLSTPLGLQTPLGIQTPLGL 126
P + R + NR+ S +A+ G+A P +A P +A GL+T P +
Sbjct 72 PRAGRSPVNGGNRQGASGKLAADGAAKP---PLAVPTVAPGLATTTAAAQPSYPSQEQRI 128
Query 127 RTPMGSSTPSLNDLGEARGTVLS 149
R + P + G G V S
Sbjct 129 RPSAHGARPGSQNCGSREGPVSS 151
> hsa:25959 KANK2, ANKRD25, DKFZp434N161, FLJ20004, KIAA1518,
MGC119707, MXRA3, SIP; KN motif and ankyrin repeat domains 2
Length=851
Score = 29.6 bits (65), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 34/80 (42%), Gaps = 10/80 (12%)
Query 65 LTPASDRGILEARNRESFSNSIASAGSATPIGFGMATPLMAGGLSTPLGLQTPLGIQTPL 124
P +R +L+AR R +ATP G G TP AG ++ +G+ P
Sbjct 120 FNPRVERTLLDARRR-------LEDQAATPTGLGSLTPSAAGSTASLVGVGLPPPTPRSS 172
Query 125 GLRTPMGSSTPSLNDLGEAR 144
GL TP+ PS L R
Sbjct 173 GLSTPV---PPSAGHLAHVR 189
> xla:733281 supt5h, spt5, spt5h, tat-ct1; suppressor of Ty 5
homolog
Length=1083
Score = 29.6 bits (65), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 23/54 (42%), Positives = 28/54 (51%), Gaps = 5/54 (9%)
Query 90 GSATPIGFGMATPLMAGGLSTPLGLQTPL--GIQTPL--GLRTPMGSSTPSLND 139
GS TP+ +G TPL G + G QTPL G +TP G P +TPS D
Sbjct 778 GSRTPM-YGSQTPLHDGSRTPHYGSQTPLHDGSRTPAQSGAWDPNNPNTPSRAD 830
> cel:T15B12.1 hypothetical protein
Length=432
Score = 29.3 bits (64), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 28/76 (36%), Positives = 38/76 (50%), Gaps = 7/76 (9%)
Query 67 PASDRGILEARNRESFSNSIASAGSATPI-GF----GMATPLMAGGLS-TPLGLQTPLGI 120
P S + E R+R S S SA+P+ GF G T + G + P G+Q PL
Sbjct 11 PQSSISLDEKRHRNR-SLGCISRYSASPLPGFSAFLGTITRVSTGQTTFHPYGVQEPLSP 69
Query 121 QTPLGLRTPMGSSTPS 136
Q+ +G+ T M STPS
Sbjct 70 QSCVGVFTSMYCSTPS 85
> mmu:269401 Znf512b, Gm632; zinc finger protein 512B
Length=869
Score = 29.3 bits (64), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 11/29 (37%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 107 GLSTPLGLQTPLGIQTPLGLRTPMGSSTP 135
G+S P+G+ P+ + P+G+ P+G S P
Sbjct 188 GVSKPIGVSKPVTVGKPVGVSKPIGISKP 216
> xla:446624 vezf1-a, MGC82370, db1, vezf1, vezf1a, znf161; vascular
endothelial zinc finger 1
Length=512
Score = 29.3 bits (64), Expect = 5.4, Method: Composition-based stats.
Identities = 14/57 (24%), Positives = 35/57 (61%), Gaps = 2/57 (3%)
Query 81 SFSNSIASAGSAT-PIGFGMATPLMAG-GLSTPLGLQTPLGIQTPLGLRTPMGSSTP 135
+ ++SIA++G+ T P+ A + + +S+P+ + +P+ + P+ + TP+ ++P
Sbjct 392 NITSSIAASGTLTNPVTVAAAMSMRSQVNVSSPVNITSPMNLGHPVTITTPISMTSP 448
> dre:557149 si:ch211-244b2.1
Length=929
Score = 29.3 bits (64), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 32/55 (58%), Gaps = 9/55 (16%)
Query 84 NSIASAGSATPIGFGMATPLMAGGLSTPLGLQT-PLGIQTPLGLRTPMGSSTPSL 137
N+I+S A P GM+T ++ G ++P+G QT +G +G+R P TP+L
Sbjct 748 NTISSTFPAAPTMGGMSTMSVSNGFNSPIGFQTGGMG----MGMRAP----TPAL 794
> hsa:6829 SUPT5H, FLJ34157, SPT5, SPT5H, Tat-CT1; suppressor
of Ty 5 homolog (S. cerevisiae)
Length=1087
Score = 29.3 bits (64), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 22/52 (42%), Positives = 28/52 (53%), Gaps = 5/52 (9%)
Query 89 AGSATPIGFGMATPLMAGGLSTPLGLQTPL--GIQTPL--GLRTPMGSSTPS 136
+GS TP+ +G TPL G + G QTPL G +TP G P +TPS
Sbjct 780 SGSRTPM-YGSQTPLQDGSRTPHYGSQTPLHDGSRTPAQSGAWDPNNPNTPS 830
> cel:W01G7.1 daf-5; abnormal DAuer Formation family member (daf-5)
Length=627
Score = 29.3 bits (64), Expect = 6.6, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 25/48 (52%), Gaps = 6/48 (12%)
Query 94 PIGFG------MATPLMAGGLSTPLGLQTPLGIQTPLGLRTPMGSSTP 135
PI G +A L+A G+ PL + TP + TP + TP+ ++ P
Sbjct 533 PISLGNINFVALAQQLIASGIKLPLPIVTPPVVSTPAPVITPIPAALP 580
> xla:779374 vezf1-b, MGC83349, db1, vezf1b, znf161; vascular
endothelial zinc finger 1
Length=511
Score = 28.9 bits (63), Expect = 6.6, Method: Composition-based stats.
Identities = 16/56 (28%), Positives = 33/56 (58%), Gaps = 6/56 (10%)
Query 81 SFSNSIASAGSAT-PIGFGMATPLMAGGLSTPLGLQTPLGIQTPLGLRTPMGSSTP 135
+ ++S+A++G+ T P+ T A + +P+ + +PL I +P+ L P+ +TP
Sbjct 391 NITSSVAASGTLTNPV-----TVAAAMSMRSPVNVSSPLNITSPMNLGHPVTITTP 441
> mmu:20924 Supt5h, AL033283, AL033321, AU019549, Spt5; suppressor
of Ty 5 homolog (S. cerevisiae)
Length=1082
Score = 28.9 bits (63), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 22/52 (42%), Positives = 28/52 (53%), Gaps = 5/52 (9%)
Query 89 AGSATPIGFGMATPLMAGGLSTPLGLQTPL--GIQTPL--GLRTPMGSSTPS 136
+GS TP+ +G TPL G + G QTPL G +TP G P +TPS
Sbjct 774 SGSRTPM-YGSQTPLQDGSRTPHYGSQTPLHDGSRTPAQSGAWDPNNPNTPS 824
> hsa:7716 VEZF1, DB1, ZNF161; vascular endothelial zinc finger
1
Length=521
Score = 28.9 bits (63), Expect = 7.6, Method: Composition-based stats.
Identities = 15/62 (24%), Positives = 34/62 (54%), Gaps = 7/62 (11%)
Query 81 SFSNSIASAGSATPI----GFGMATPL---MAGGLSTPLGLQTPLGIQTPLGLRTPMGSS 133
S ++S++S + P+ M +P+ A +++P+ + P+ I +PL + +P+ +
Sbjct 402 SITSSVSSGTMSNPVTVAAAMSMRSPVNVSSAVNITSPMNIGHPVTITSPLSMTSPLTLT 461
Query 134 TP 135
TP
Sbjct 462 TP 463
Lambda K H
0.312 0.130 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3712313100
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40