bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1271_orf2
Length=130
Score E
Sequences producing significant alignments: (Bits) Value
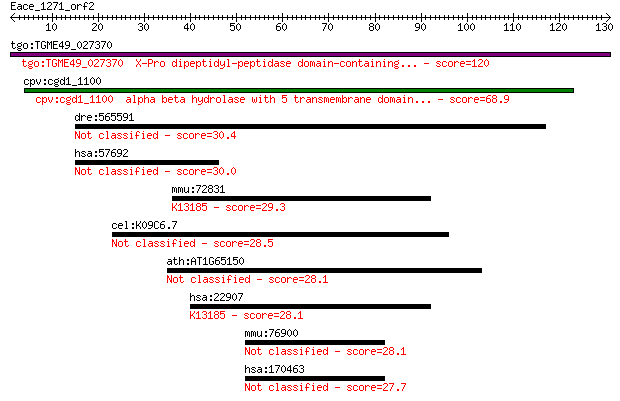
tgo:TGME49_027370 X-Pro dipeptidyl-peptidase domain-containing... 120 1e-27
cpv:cgd1_1100 alpha beta hydrolase with 5 transmembrane domain... 68.9 4e-12
dre:565591 FAT tumor suppressor homolog 1-like 30.4 1.5
hsa:57692 MAGEE1, DAMAGE, HCA1, KIAA1587; melanoma antigen fam... 30.0 1.9
mmu:72831 Dhx30, 2810477H02Rik, C130058C04Rik, Ddx30, HELG, Re... 29.3 2.9
cel:K09C6.7 hypothetical protein 28.5 5.2
ath:AT1G65150 meprin and TRAF homology domain-containing prote... 28.1 6.6
hsa:22907 DHX30, DDX30, FLJ11214, KIAA0890, RETCOR; DEAH (Asp-... 28.1 7.6
mmu:76900 Ssbp4, 1210002E11Rik, AW743380, Sspb4; single strand... 28.1 8.0
hsa:170463 SSBP4, MGC3181; single stranded DNA binding protein 4 27.7
> tgo:TGME49_027370 X-Pro dipeptidyl-peptidase domain-containing
protein (EC:3.4.14.11); K06978
Length=1177
Score = 120 bits (301), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 56/131 (42%), Positives = 88/131 (67%), Gaps = 1/131 (0%)
Query 1 LERDLQMVGSAFLNLFIKAKHCNEISLFAYLEDVDVAAGFSHYVTEAQVLASNRPSEVRD 60
L ++ +VGSA+++L + K C +I++F YLE VDV G+SHYVTE +++ S+RP V++
Sbjct 1024 LVEEVDVVGSAWIHLQMFVKSCTDIAVFVYLEAVDVNGGYSHYVTEGKIVVSHRPVTVKE 1083
Query 61 DLPVGSYGRVLRPFSRKSYKPIEEDEE-VNVSVNFEPTAWTFKKGHSLRLLLTGSDLENF 119
+ P+GS V RPF++ S P+ E + ++ EP +WTFKKG ++RL L GSD++NF
Sbjct 1084 ESPIGSPDTVFRPFTKSSRIPLNSSGELIEFTMPLEPVSWTFKKGQAIRLSLGGSDVDNF 1143
Query 120 QLNPAKEVLLP 130
Q ++LP
Sbjct 1144 QPAVQNRMILP 1154
> cpv:cgd1_1100 alpha beta hydrolase with 5 transmembrane domains
; K06978
Length=1103
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 37/124 (29%), Positives = 67/124 (54%), Gaps = 6/124 (4%)
Query 4 DLQMVGSAFLNLFIKAKHCNEISLFAYLEDVDVAAGFSHYVTEAQVLASNR----PSEVR 59
D+++VGS +L+ ++ + ++ YLED+ Y+TE +R
Sbjct 940 DIRLVGSVWLHFSVQLLDGYDAPIYVYLEDISPENEI-MYITEGLAQIGHRHVNMTKSAL 998
Query 60 DDLPVGSYGRVLRPFSRKSYKPIEE-DEEVNVSVNFEPTAWTFKKGHSLRLLLTGSDLEN 118
+++P GS +V+RP +R Y+P+ +E + + EP WTF K H L + +TG+D +N
Sbjct 999 ENIPHGSARKVIRPLTRSYYQPLRRIGDESPMQITLEPITWTFFKDHRLAISITGADTKN 1058
Query 119 FQLN 122
F+L+
Sbjct 1059 FKLH 1062
> dre:565591 FAT tumor suppressor homolog 1-like
Length=4472
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 30/110 (27%), Positives = 56/110 (50%), Gaps = 10/110 (9%)
Query 15 LFIKAKHCNEISLFAYLEDVDVAAGFSHYVTEAQVLASNRPSEVRDDLPVGSYGRVLRPF 74
L IKA E SL A + V + S T+ + +++ +EV +++PVGS+ ++ F
Sbjct 1629 LIIKASDKGEPSLSA-VTAVRITVTISDN-TKPKFPSNSLSAEVSENVPVGSFVTLVSAF 1686
Query 75 SRKS--YKPIEEDEEVNVSVNFEPTAWTFKKG------HSLRLLLTGSDL 116
S+ + Y+ IE +E+ +N A +K S +L++ G+D+
Sbjct 1687 SQSTVFYQIIEGNEKGAFDINPNSGAVITRKKLDFETLSSYKLIVQGTDM 1736
> hsa:57692 MAGEE1, DAMAGE, HCA1, KIAA1587; melanoma antigen family
E, 1
Length=957
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 11/31 (35%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 15 LFIKAKHCNEISLFAYLEDVDVAAGFSHYVT 45
+FI H E +++A+L + V AG H +T
Sbjct 845 IFIMGNHARESAVWAFLRGLGVQAGRKHVIT 875
> mmu:72831 Dhx30, 2810477H02Rik, C130058C04Rik, Ddx30, HELG,
Ret-CoR; DEAH (Asp-Glu-Ala-His) box polypeptide 30 (EC:3.6.4.13);
K13185 ATP-dependent RNA helicase DHX30 [EC:3.6.4.13]
Length=1217
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 29/57 (50%), Gaps = 1/57 (1%)
Query 36 VAAGFSHYVTEAQVLASNRPSEVRDDLPVGS-YGRVLRPFSRKSYKPIEEDEEVNVS 91
+ A SHY ++ ++ + D LP+G G + P + K Y P+ E EEV +S
Sbjct 381 IEAFLSHYPVDSSWISPELRLQSDDILPLGKDSGPLSDPITGKPYMPLSEAEEVRLS 437
> cel:K09C6.7 hypothetical protein
Length=587
Score = 28.5 bits (62), Expect = 5.2, Method: Composition-based stats.
Identities = 17/74 (22%), Positives = 33/74 (44%), Gaps = 2/74 (2%)
Query 23 NEISLFAYLEDVDVAAGFSHYVTEAQVLASNRPSEVRDDLPVGSYGRVL-RPFSRKSYKP 81
N+ + L V++ + F H + + N P +DD+ +G+ GRV+ R + Y
Sbjct 225 NKFPVVQTLNSVEIESNFKHISKGTKFMLGNMPMTTKDDV-IGNMGRVVCRYLALNQYHQ 283
Query 82 IEEDEEVNVSVNFE 95
++NF+
Sbjct 284 TIYHFHFRAAINFD 297
> ath:AT1G65150 meprin and TRAF homology domain-containing protein
/ MATH domain-containing protein
Length=296
Score = 28.1 bits (61), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 20/72 (27%), Positives = 33/72 (45%), Gaps = 9/72 (12%)
Query 35 DVAAGFSHYVTE----AQVLASNRPSEVRDDLPVGSYGRVLRPFSRKSYKPIEEDEEVNV 90
D A GF Y E A V +RP+ +DLP + +R F+ +E+++ V+
Sbjct 127 DRAKGFILYGEEHEFGAHVKIVSRPASFGEDLPFHKFSWTIRDFAL-----LEQNDYVSK 181
Query 91 SVNFEPTAWTFK 102
+ + WT K
Sbjct 182 TFHMGEKDWTLK 193
> hsa:22907 DHX30, DDX30, FLJ11214, KIAA0890, RETCOR; DEAH (Asp-Glu-Ala-His)
box polypeptide 30 (EC:3.6.4.13); K13185 ATP-dependent
RNA helicase DHX30 [EC:3.6.4.13]
Length=1155
Score = 28.1 bits (61), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 27/53 (50%), Gaps = 1/53 (1%)
Query 40 FSHYVTEAQVLASNRPSEVRDDLPVGS-YGRVLRPFSRKSYKPIEEDEEVNVS 91
+HY E+ +A + D LP+G G + P + K Y P+ E EEV +S
Sbjct 323 LNHYPVESSWIAPELRLQSDDILPLGKDSGPLSDPITGKPYVPLLEAEEVRLS 375
> mmu:76900 Ssbp4, 1210002E11Rik, AW743380, Sspb4; single stranded
DNA binding protein 4
Length=363
Score = 28.1 bits (61), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 16/30 (53%), Gaps = 0/30 (0%)
Query 52 SNRPSEVRDDLPVGSYGRVLRPFSRKSYKP 81
SN P RDD + + G L PF +SY P
Sbjct 328 SNAPGTPRDDGEMAAAGTFLHPFPSESYSP 357
> hsa:170463 SSBP4, MGC3181; single stranded DNA binding protein
4
Length=363
Score = 27.7 bits (60), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 16/30 (53%), Gaps = 0/30 (0%)
Query 52 SNRPSEVRDDLPVGSYGRVLRPFSRKSYKP 81
SN P RDD + + G L PF +SY P
Sbjct 328 SNAPGTPRDDGEMAAAGTFLHPFPSESYSP 357
Lambda K H
0.318 0.135 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2039374804
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40