bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1119_orf1
Length=150
Score E
Sequences producing significant alignments: (Bits) Value
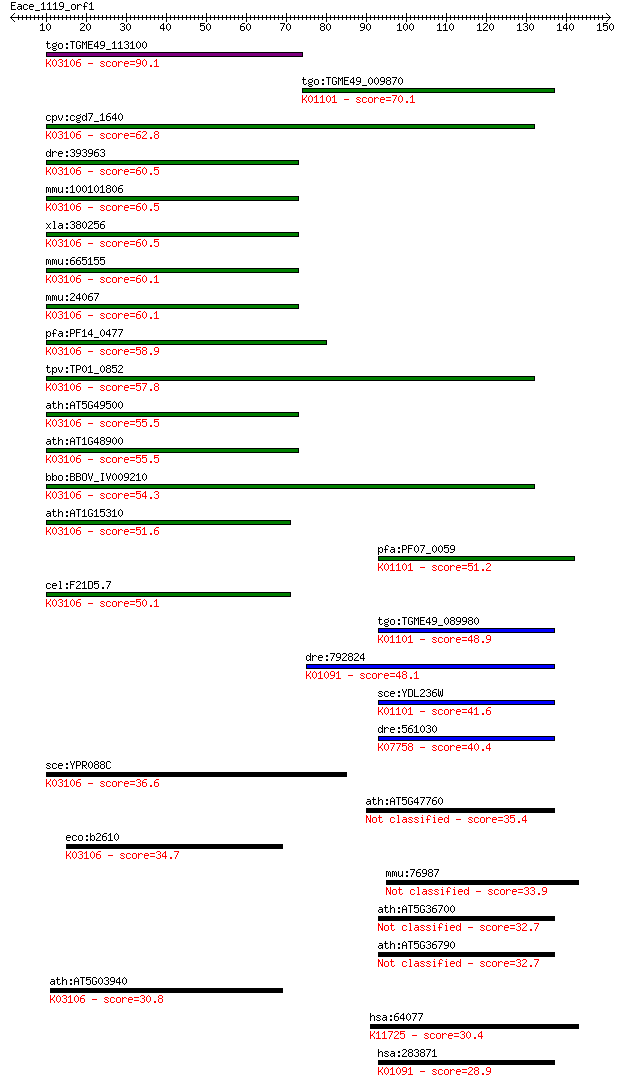
tgo:TGME49_113100 signal recognition particle 54 kda protein, ... 90.1 2e-18
tgo:TGME49_009870 pyridoxal phosphate phosphatase, putative (E... 70.1 3e-12
cpv:cgd7_1640 SRP54. signal recognition 54. GTpase. ; K03106 s... 62.8 4e-10
dre:393963 srp54, MGC55842, Srp54k, zgc:55842, zgc:85713; sign... 60.5 2e-09
mmu:100101806 Srp54c, E330029E12Rik, MGC6231; signal recogniti... 60.5 2e-09
xla:380256 srp54, MGC53033; signal recognition particle 54kDa;... 60.5 2e-09
mmu:665155 Srp54b; signal recognition particle 54B; K03106 sig... 60.1 3e-09
mmu:24067 Srp54a, 54kDa, Srp54; signal recognition particle 54... 60.1 3e-09
pfa:PF14_0477 signal recognition particle SRP54, putative; K03... 58.9 5e-09
tpv:TP01_0852 signal recognition particle 54 kDa protein; K031... 57.8 1e-08
ath:AT5G49500 signal recognition particle 54 kDa protein 2 / S... 55.5 6e-08
ath:AT1G48900 signal recognition particle 54 kDa protein 3 / S... 55.5 7e-08
bbo:BBOV_IV009210 23.m06227; signal recognition particle SRP54... 54.3 1e-07
ath:AT1G15310 ATHSRP54A (ARABIDOPSIS THALIANA SIGNAL RECOGNITI... 51.6 8e-07
pfa:PF07_0059 4-nitrophenylphosphatase, putative (EC:3.1.3.41)... 51.2 1e-06
cel:F21D5.7 hypothetical protein; K03106 signal recognition pa... 50.1 3e-06
tgo:TGME49_089980 4-nitrophenylphosphatase, putative (EC:3.1.3... 48.9 6e-06
dre:792824 pgp, MGC56011, cb241, sb:cb241, wu:fb92c03, wu:fi40... 48.1 1e-05
sce:YDL236W PHO13; Alkaline phosphatase specific for p-nitroph... 41.6 0.001
dre:561030 zgc:194409 (EC:3.1.3.74); K07758 pyridoxal phosphat... 40.4 0.002
sce:YPR088C SRP54, SRH1; Signal recognition particle (SRP) sub... 36.6 0.032
ath:AT5G47760 ATPGLP2 (ARABIDOPSIS THALIANA 2-PHOSPHOGLYCOLATE... 35.4 0.069
eco:b2610 ffh, ECK2607, JW5414; signal recognition particle (S... 34.7 0.12
mmu:76987 Hdhd2, 0610039H12Rik, 3110052N05Rik; haloacid dehalo... 33.9 0.20
ath:AT5G36700 PGLP1; PGLP1 (2-PHOSPHOGLYCOLATE PHOSPHATASE 1);... 32.7 0.46
ath:AT5G36790 phosphoglycolate phosphatase, putative (EC:3.1.3... 32.7 0.46
ath:AT5G03940 CPSRP54 (CHLOROPLAST SIGNAL RECOGNITION PARTICLE... 30.8 1.9
hsa:64077 LHPP, FLJ44846, FLJ46044, HDHD2B, MGC117251, MGC1421... 30.4 2.0
hsa:283871 PGP, MGC4692; phosphoglycolate phosphatase (EC:3.1.... 28.9 7.2
> tgo:TGME49_113100 signal recognition particle 54 kda protein,
putative (EC:2.7.1.25); K03106 signal recognition particle
subunit SRP54
Length=582
Score = 90.1 bits (222), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 43/64 (67%), Positives = 51/64 (79%), Gaps = 0/64 (0%)
Query 10 MVLAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQI 69
MVLAELG+QISGALR L +AT+VDE VV +CV VCRALL ADV +RVVQ+FK VT I
Sbjct 1 MVLAELGEQISGALRRLHTATVVDEQVVQECVMAVCRALLQADVHVRVVQQFKASVTAGI 60
Query 70 NTQI 73
N ++
Sbjct 61 NNRL 64
> tgo:TGME49_009870 pyridoxal phosphate phosphatase, putative
(EC:3.1.3.41); K01101 4-nitrophenyl phosphatase [EC:3.1.3.41]
Length=491
Score = 70.1 bits (170), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 30/63 (47%), Positives = 45/63 (71%), Gaps = 0/63 (0%)
Query 74 RQQSSNSSSSSSSSSSSSKKSVYFLTNNSTLSRKGFVAKLNKLGIEATEQQVLCTSYSTA 133
R+ ++ SS+S S + +K +YFLTNNST SR+GF+ KL LG+ ATE+QV+C+S +
Sbjct 96 RELPDDAGRSSASGSGAQQKKIYFLTNNSTKSRRGFLKKLESLGVHATEEQVVCSSVVAS 155
Query 134 RYI 136
Y+
Sbjct 156 WYL 158
> cpv:cgd7_1640 SRP54. signal recognition 54. GTpase. ; K03106
signal recognition particle subunit SRP54
Length=515
Score = 62.8 bits (151), Expect = 4e-10, Method: Composition-based stats.
Identities = 38/128 (29%), Positives = 68/128 (53%), Gaps = 12/128 (9%)
Query 10 MVLAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQI 69
MVLA+LG Q++ A+R QS+TI DEA ++ C+K + ALL ADV +++V + + + + +
Sbjct 16 MVLADLGGQLASAIRKFQSSTIADEAAIDLCLKEIATALLKADVNVKLVAQLRNNIKKAV 75
Query 70 NTQIRQQSSNSSSSSSSSSSSSKKSVYFLTNNSTLSRKGFVAKLNK------LGIEATEQ 123
+ +S S+ V L N T SR +V + +K +G++ + +
Sbjct 76 ------EQEDSVGGSNRRRQIQSAVVEELVNILTPSRTPYVPQRDKSNVIVFVGLQGSGK 129
Query 124 QVLCTSYS 131
CT ++
Sbjct 130 TTTCTKFA 137
> dre:393963 srp54, MGC55842, Srp54k, zgc:55842, zgc:85713; signal
recognition particle 54; K03106 signal recognition particle
subunit SRP54
Length=504
Score = 60.5 bits (145), Expect = 2e-09, Method: Composition-based stats.
Identities = 29/63 (46%), Positives = 47/63 (74%), Gaps = 0/63 (0%)
Query 10 MVLAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQI 69
MVLA+LG++I+ ALR+L +ATI++E V+N +K VC ALL ADV +++V++ + V I
Sbjct 1 MVLADLGRKITSALRSLSNATIINEEVLNAMLKEVCAALLEADVNIKLVKQLRENVKAAI 60
Query 70 NTQ 72
+ +
Sbjct 61 DLE 63
> mmu:100101806 Srp54c, E330029E12Rik, MGC6231; signal recognition
particle 54C; K03106 signal recognition particle subunit
SRP54
Length=504
Score = 60.5 bits (145), Expect = 2e-09, Method: Composition-based stats.
Identities = 29/63 (46%), Positives = 47/63 (74%), Gaps = 0/63 (0%)
Query 10 MVLAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQI 69
MVLA+LG++I+ ALR+L +ATI++E V+N +K VC ALL ADV +++V++ + V I
Sbjct 1 MVLADLGRKITSALRSLSNATIINEEVLNAMLKEVCTALLEADVNIKLVKQLRENVKSAI 60
Query 70 NTQ 72
+ +
Sbjct 61 DLE 63
> xla:380256 srp54, MGC53033; signal recognition particle 54kDa;
K03106 signal recognition particle subunit SRP54
Length=504
Score = 60.5 bits (145), Expect = 2e-09, Method: Composition-based stats.
Identities = 29/63 (46%), Positives = 47/63 (74%), Gaps = 0/63 (0%)
Query 10 MVLAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQI 69
MVLA+LG++I+ ALR+L +ATI++E V+N +K VC ALL ADV +++V++ + V I
Sbjct 1 MVLADLGRKITSALRSLSNATIINEEVLNAMLKEVCTALLEADVNIKLVKQLRENVKSAI 60
Query 70 NTQ 72
+ +
Sbjct 61 DLE 63
> mmu:665155 Srp54b; signal recognition particle 54B; K03106 signal
recognition particle subunit SRP54
Length=504
Score = 60.1 bits (144), Expect = 3e-09, Method: Composition-based stats.
Identities = 29/63 (46%), Positives = 47/63 (74%), Gaps = 0/63 (0%)
Query 10 MVLAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQI 69
MVLA+LG++I+ ALR+L +ATI++E V+N +K VC ALL ADV +++V++ + V I
Sbjct 1 MVLADLGRKITSALRSLSNATIINEEVLNAMLKEVCTALLEADVNIKLVKQLRENVKSAI 60
Query 70 NTQ 72
+ +
Sbjct 61 DLE 63
> mmu:24067 Srp54a, 54kDa, Srp54; signal recognition particle
54A; K03106 signal recognition particle subunit SRP54
Length=504
Score = 60.1 bits (144), Expect = 3e-09, Method: Composition-based stats.
Identities = 29/63 (46%), Positives = 47/63 (74%), Gaps = 0/63 (0%)
Query 10 MVLAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQI 69
MVLA+LG++I+ ALR+L +ATI++E V+N +K VC ALL ADV +++V++ + V I
Sbjct 1 MVLADLGRKITSALRSLSNATIINEEVLNAMLKEVCTALLEADVNIKLVKQLRENVKSAI 60
Query 70 NTQ 72
+ +
Sbjct 61 DLE 63
> pfa:PF14_0477 signal recognition particle SRP54, putative; K03106
signal recognition particle subunit SRP54
Length=500
Score = 58.9 bits (141), Expect = 5e-09, Method: Composition-based stats.
Identities = 26/70 (37%), Positives = 44/70 (62%), Gaps = 0/70 (0%)
Query 10 MVLAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQI 69
MVL ELG QI+ A R LQ++T+ D+ V+ +C+K + RAL+L+D+ + +++ K + I
Sbjct 1 MVLTELGTQITNAFRKLQTSTLADDVVIEECLKEIIRALILSDINVSYLKDIKSNIKNNI 60
Query 70 NTQIRQQSSN 79
I +N
Sbjct 61 EKNIDIYGNN 70
> tpv:TP01_0852 signal recognition particle 54 kDa protein; K03106
signal recognition particle subunit SRP54
Length=495
Score = 57.8 bits (138), Expect = 1e-08, Method: Composition-based stats.
Identities = 39/129 (30%), Positives = 64/129 (49%), Gaps = 14/129 (10%)
Query 10 MVLAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQI 69
MVLAEL QI+ A R L S T++ EAV+ + + + RALL+ADV +++V + K V
Sbjct 1 MVLAELSNQITKAFRKLHSTTVISEAVIEEVIGDIVRALLMADVNVKLVHKLKENVK--- 57
Query 70 NTQIRQQSSNSSSSSSSSSSSSKKSVY-FLTNNSTLSRKGFVAKLNK------LGIEATE 122
R +N+ ++ +K V L N T +K F K + +G++
Sbjct 58 ----RLNKNNADMIGANKRRYLQKIVVDELVNMLTTEKKPFEPKKGRCNVIMFVGLQGAG 113
Query 123 QQVLCTSYS 131
+ CT ++
Sbjct 114 KTTSCTKFA 122
> ath:AT5G49500 signal recognition particle 54 kDa protein 2 /
SRP54 (SRP-54B); K03106 signal recognition particle subunit
SRP54
Length=497
Score = 55.5 bits (132), Expect = 6e-08, Method: Composition-based stats.
Identities = 25/63 (39%), Positives = 42/63 (66%), Gaps = 0/63 (0%)
Query 10 MVLAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQI 69
MVLAELG +I A++ + + TI+DE +NDC+ + RALL +DV +V+E + + + +
Sbjct 1 MVLAELGGRIMSAIQKMNNVTIIDEKALNDCLNEITRALLQSDVSFPLVKEMQTNIKKIV 60
Query 70 NTQ 72
N +
Sbjct 61 NLE 63
> ath:AT1G48900 signal recognition particle 54 kDa protein 3 /
SRP54 (SRP-54C); K03106 signal recognition particle subunit
SRP54
Length=431
Score = 55.5 bits (132), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 24/63 (38%), Positives = 43/63 (68%), Gaps = 0/63 (0%)
Query 10 MVLAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQI 69
MVLAELG +I+ A++ + + TI+DE +N+C+ + RALL +DV +V+E + + + +
Sbjct 1 MVLAELGGRITRAIQQMSNVTIIDEKALNECLNEITRALLQSDVSFPLVKEMQSNIKKIV 60
Query 70 NTQ 72
N +
Sbjct 61 NLE 63
> bbo:BBOV_IV009210 23.m06227; signal recognition particle SRP54
protein; K03106 signal recognition particle subunit SRP54
Length=499
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 31/122 (25%), Positives = 60/122 (49%), Gaps = 0/122 (0%)
Query 10 MVLAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQI 69
MVLAEL +QI+ A R L + TI+ E V+++ + + RALL ADV +++V++ + V Q+
Sbjct 1 MVLAELSEQITQAFRKLHARTIITEEVIDEVIGDIVRALLSADVNVKLVRQLRENVKLQV 60
Query 70 NTQIRQQSSNSSSSSSSSSSSSKKSVYFLTNNSTLSRKGFVAKLNKLGIEATEQQVLCTS 129
+N + ++ + +KG + +G++ + CT
Sbjct 61 RLHADALGANKRKFIQKAVIEEFINMLSSDRKPYVPKKGHANIIMFVGLQGAGKTTTCTK 120
Query 130 YS 131
++
Sbjct 121 FA 122
> ath:AT1G15310 ATHSRP54A (ARABIDOPSIS THALIANA SIGNAL RECOGNITION
PARTICLE 54 KDA SUBUNIT); 7S RNA binding / GTP binding
/ mRNA binding / nucleoside-triphosphatase/ nucleotide binding;
K03106 signal recognition particle subunit SRP54
Length=479
Score = 51.6 bits (122), Expect = 8e-07, Method: Composition-based stats.
Identities = 24/61 (39%), Positives = 42/61 (68%), Gaps = 0/61 (0%)
Query 10 MVLAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQI 69
MVLAELG +I+ A++ + + TI+DE V+ND + + RALL +DV +V++ + + + +
Sbjct 1 MVLAELGGRITRAIQQMNNVTIIDEKVLNDFLNEITRALLQSDVSFGLVEKMQTNIKKIV 60
Query 70 N 70
N
Sbjct 61 N 61
> pfa:PF07_0059 4-nitrophenylphosphatase, putative (EC:3.1.3.41);
K01101 4-nitrophenyl phosphatase [EC:3.1.3.41]
Length=322
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 20/50 (40%), Positives = 33/50 (66%), Gaps = 1/50 (2%)
Query 93 KSVYFLTNNSTLSRKGFVAKLNKLGI-EATEQQVLCTSYSTARYILSQQQ 141
K VYF+TNNST SR F+ K +KLG + ++CT+Y+ +Y+ +++
Sbjct 76 KKVYFITNNSTKSRASFLEKFHKLGFTNVKREHIICTAYAVTKYLYDKEE 125
> cel:F21D5.7 hypothetical protein; K03106 signal recognition
particle subunit SRP54
Length=496
Score = 50.1 bits (118), Expect = 3e-06, Method: Composition-based stats.
Identities = 24/61 (39%), Positives = 42/61 (68%), Gaps = 0/61 (0%)
Query 10 MVLAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQI 69
MVLA+LG++I A+ L +T+++E ++ +K VC AL+ +DV +R+V++ K V + I
Sbjct 1 MVLADLGRKIRNAIGKLGQSTVINEGELDLMLKEVCTALIESDVHIRLVKQLKDNVKKAI 60
Query 70 N 70
N
Sbjct 61 N 61
> tgo:TGME49_089980 4-nitrophenylphosphatase, putative (EC:3.1.3.41
2.7.7.7); K01101 4-nitrophenyl phosphatase [EC:3.1.3.41]
Length=593
Score = 48.9 bits (115), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 21/44 (47%), Positives = 30/44 (68%), Gaps = 0/44 (0%)
Query 93 KSVYFLTNNSTLSRKGFVAKLNKLGIEATEQQVLCTSYSTARYI 136
K V F TN ++ SR+ VA L K G EA E++++CTSY+ A Y+
Sbjct 314 KRVIFFTNGASKSRRTCVALLRKAGFEAHEEEMICTSYAAAEYM 357
> dre:792824 pgp, MGC56011, cb241, sb:cb241, wu:fb92c03, wu:fi40b02,
zgc:56011; phosphoglycolate phosphatase (EC:3.1.3.18);
K01091 phosphoglycolate phosphatase [EC:3.1.3.18]
Length=306
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 36/62 (58%), Gaps = 0/62 (0%)
Query 75 QQSSNSSSSSSSSSSSSKKSVYFLTNNSTLSRKGFVAKLNKLGIEATEQQVLCTSYSTAR 134
Q+ + +S K V+F+TNNST +R+ + KL KLG +A +V T+Y +A+
Sbjct 36 DQAIPGAPEVINSLKKHGKQVFFVTNNSTKTRQMYADKLGKLGFDAAADEVFGTAYCSAQ 95
Query 135 YI 136
Y+
Sbjct 96 YL 97
> sce:YDL236W PHO13; Alkaline phosphatase specific for p-nitrophenyl
phosphate; also has protein phosphatase activity (EC:3.1.3.41);
K01101 4-nitrophenyl phosphatase [EC:3.1.3.41]
Length=312
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 27/44 (61%), Gaps = 0/44 (0%)
Query 93 KSVYFLTNNSTLSRKGFVAKLNKLGIEATEQQVLCTSYSTARYI 136
K + F+TNNST SR + K GI+ E+Q+ + Y++A YI
Sbjct 57 KQLIFVTNNSTKSRLAYTKKFASFGIDVKEEQIFTSGYASAVYI 100
> dre:561030 zgc:194409 (EC:3.1.3.74); K07758 pyridoxal phosphatase
[EC:3.1.3.74]
Length=308
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 30/45 (66%), Gaps = 1/45 (2%)
Query 93 KSVYFLTNNSTLSRKGFVAKLNKLGI-EATEQQVLCTSYSTARYI 136
K V+F+TNN T R+ +V K ++LG + E+++ ++Y +A Y+
Sbjct 56 KRVFFVTNNCTRPRENYVQKFSRLGFADVAEEEIFSSAYCSAAYL 100
> sce:YPR088C SRP54, SRH1; Signal recognition particle (SRP) subunit
(homolog of mammalian SRP54); contains the signal sequence-binding
activity of SRP, interacts with the SRP RNA, and
mediates binding of SRP to signal receptor; contains GTPase
domain; K03106 signal recognition particle subunit SRP54
Length=541
Score = 36.6 bits (83), Expect = 0.032, Method: Composition-based stats.
Identities = 23/76 (30%), Positives = 45/76 (59%), Gaps = 1/76 (1%)
Query 10 MVLAELGQQISGALRNLQSATIVD-EAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQ 68
MVLA+LG++I+ A+ N S T D V+ +K + ALL +DV + +V + + + Q
Sbjct 1 MVLADLGKRINSAVNNAISNTQDDFTTSVDVMLKGIVTALLESDVNIALVSKLRNNIRSQ 60
Query 69 INTQIRQQSSNSSSSS 84
+ ++ R + S +++ +
Sbjct 61 LLSENRSEKSTTNAQT 76
> ath:AT5G47760 ATPGLP2 (ARABIDOPSIS THALIANA 2-PHOSPHOGLYCOLATE
PHOSPHATASE 2); phosphoglycolate phosphatase/ protein serine/threonine
kinase (EC:3.1.3.18)
Length=301
Score = 35.4 bits (80), Expect = 0.069, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 30/48 (62%), Gaps = 1/48 (2%)
Query 90 SSKKSVYFLTNNSTLSRKGFVAKLNKLGIEA-TEQQVLCTSYSTARYI 136
S K+V F+TNNS SR+ + K LG+ + T+ ++ +S++ A Y+
Sbjct 48 SKGKNVVFVTNNSVKSRRQYAEKFRSLGVTSITQDEIFSSSFAAAMYL 95
> eco:b2610 ffh, ECK2607, JW5414; signal recognition particle
(SRP) component with 4.5S RNA (ffs); K03106 signal recognition
particle subunit SRP54
Length=453
Score = 34.7 bits (78), Expect = 0.12, Method: Composition-based stats.
Identities = 21/54 (38%), Positives = 31/54 (57%), Gaps = 0/54 (0%)
Query 15 LGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQ 68
L ++S LRN+ + E V D ++ V ALL ADV L VV+EF +V ++
Sbjct 5 LTDRLSRTLRNISGRGRLTEDNVKDTLREVRMALLEADVALPVVREFINRVKEK 58
> mmu:76987 Hdhd2, 0610039H12Rik, 3110052N05Rik; haloacid dehalogenase-like
hydrolase domain containing 2
Length=259
Score = 33.9 bits (76), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 30/48 (62%), Gaps = 1/48 (2%)
Query 95 VYFLTNNSTLSRKGFVAKLNKLGIEATEQQVLCTSYSTARYILSQQQL 142
V F+TN + S+K + +L KL E +E ++ TS + AR ++ Q+Q+
Sbjct 42 VRFVTNTTKESKKDLLERLKKLEFEISEDEIF-TSLTAARNLIEQKQV 88
> ath:AT5G36700 PGLP1; PGLP1 (2-PHOSPHOGLYCOLATE PHOSPHATASE 1);
phosphoglycolate phosphatase (EC:3.1.3.18)
Length=362
Score = 32.7 bits (73), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 28/44 (63%), Gaps = 0/44 (0%)
Query 93 KSVYFLTNNSTLSRKGFVAKLNKLGIEATEQQVLCTSYSTARYI 136
K + F+TNNST SRK + K LG+ E+++ +S++ A Y+
Sbjct 112 KRLVFVTNNSTKSRKQYGKKFETLGLNVNEEEIFASSFAAAAYL 155
> ath:AT5G36790 phosphoglycolate phosphatase, putative (EC:3.1.3.18)
Length=362
Score = 32.7 bits (73), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 28/44 (63%), Gaps = 0/44 (0%)
Query 93 KSVYFLTNNSTLSRKGFVAKLNKLGIEATEQQVLCTSYSTARYI 136
K + F+TNNST SRK + K LG+ E+++ +S++ A Y+
Sbjct 112 KRLVFVTNNSTKSRKQYGKKFETLGLNVNEEEIFASSFAAAAYL 155
> ath:AT5G03940 CPSRP54 (CHLOROPLAST SIGNAL RECOGNITION PARTICLE
54 KDA SUBUNIT); 7S RNA binding / GTP binding / mRNA binding
/ signal sequence binding; K03106 signal recognition particle
subunit SRP54
Length=564
Score = 30.8 bits (68), Expect = 1.9, Method: Composition-based stats.
Identities = 22/62 (35%), Positives = 33/62 (53%), Gaps = 4/62 (6%)
Query 11 VLAELGQQISGALRNLQSATIVDEAVVNDCV----KTVCRALLLADVQLRVVQEFKRKVT 66
V AE+ Q++G L S +E + D + + + RALL ADV L VV+ F + V+
Sbjct 73 VRAEMFGQLTGGLEAAWSKLKGEEVLTKDNIAEPMRDIRRALLEADVSLPVVRRFVQSVS 132
Query 67 QQ 68
Q
Sbjct 133 DQ 134
> hsa:64077 LHPP, FLJ44846, FLJ46044, HDHD2B, MGC117251, MGC142189,
MGC142191; phospholysine phosphohistidine inorganic pyrophosphate
phosphatase (EC:3.6.1.1); K11725 phospholysine phosphohistidine
inorganic pyrophosphate phosphatase [EC:3.6.1.1
3.1.3.-]
Length=210
Score = 30.4 bits (67), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 29/52 (55%), Gaps = 1/52 (1%)
Query 91 SKKSVYFLTNNSTLSRKGFVAKLNKLGIEATEQQVLCTSYSTARYILSQQQL 142
S+ V F TN S SR V +L +LG + +EQ+V + + + IL +Q L
Sbjct 46 SRLKVRFCTNESQKSRAELVGQLQRLGFDISEQEVTAPAPAACQ-ILKEQGL 96
> hsa:283871 PGP, MGC4692; phosphoglycolate phosphatase (EC:3.1.3.18);
K01091 phosphoglycolate phosphatase [EC:3.1.3.18]
Length=321
Score = 28.9 bits (63), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 26/49 (53%), Gaps = 5/49 (10%)
Query 93 KSVYFLTNNSTLSRKGFVAKLNKLGIEATEQ-----QVLCTSYSTARYI 136
K + F+TNNS+ +R + KL +LG +V T+Y TA Y+
Sbjct 61 KRLGFITNNSSKTRAAYAEKLRRLGFGGPAGPGASLEVFGTAYCTALYL 109
Lambda K H
0.312 0.120 0.307
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3134054208
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40