bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1111_orf2
Length=262
Score E
Sequences producing significant alignments: (Bits) Value
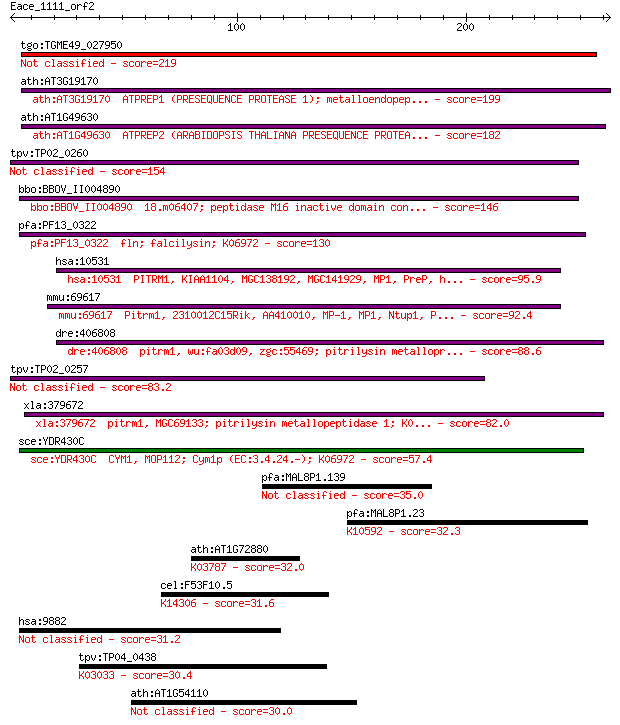
tgo:TGME49_027950 zinc metalloprotease 2, putative (EC:3.4.24.55) 219 7e-57
ath:AT3G19170 ATPREP1 (PRESEQUENCE PROTEASE 1); metalloendopep... 199 8e-51
ath:AT1G49630 ATPREP2 (ARABIDOPSIS THALIANA PRESEQUENCE PROTEA... 182 1e-45
tpv:TP02_0260 falcilysin 154 2e-37
bbo:BBOV_II004890 18.m06407; peptidase M16 inactive domain con... 146 9e-35
pfa:PF13_0322 fln; falcilysin; K06972 130 6e-30
hsa:10531 PITRM1, KIAA1104, MGC138192, MGC141929, MP1, PreP, h... 95.9 1e-19
mmu:69617 Pitrm1, 2310012C15Rik, AA410010, MP-1, MP1, Ntup1, P... 92.4 2e-18
dre:406808 pitrm1, wu:fa03d09, zgc:55469; pitrilysin metallopr... 88.6 2e-17
tpv:TP02_0257 falcilysin 83.2 1e-15
xla:379672 pitrm1, MGC69133; pitrilysin metallopeptidase 1; K0... 82.0 2e-15
sce:YDR430C CYM1, MOP112; Cym1p (EC:3.4.24.-); K06972 57.4 5e-08
pfa:MAL8P1.139 conserved Plasmodium membrane protein, unknown ... 35.0 0.29
pfa:MAL8P1.23 ubiquitin-protein ligase 1, putative; K10592 E3 ... 32.3 1.9
ath:AT1G72880 acid phosphatase survival protein SurE, putative... 32.0 2.2
cel:F53F10.5 npp-11; Nuclear Pore complex Protein family membe... 31.6 3.3
hsa:9882 TBC1D4, AS160, DKFZp779C0666; TBC1 domain family, mem... 31.2 4.4
tpv:TP04_0438 proteasome regulatory component; K03033 26S prot... 30.4 6.8
ath:AT1G54110 cation exchanger, putative (CAX10) 30.0 7.8
> tgo:TGME49_027950 zinc metalloprotease 2, putative (EC:3.4.24.55)
Length=1728
Score = 219 bits (558), Expect = 7e-57, Method: Compositional matrix adjust.
Identities = 118/255 (46%), Positives = 164/255 (64%), Gaps = 10/255 (3%)
Query 6 RMALQLLSYLLLGTSSSPLYKALSESGLGKSVIGGGLGLDLRHATFSVGLKGVAQE--ED 63
R L +L++LL+GTS SPLY+AL+ESGLGK V+G L L+H F+ GLKG+ Q+ ED
Sbjct 956 RQTLGVLTHLLVGTSPSPLYRALTESGLGKQVMGE-LEDGLKHLIFTAGLKGIPQQSAED 1014
Query 64 --MVEKVEKVIFRCLSSLSEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSIILDMATE 121
+V+KVE+++ CL + EGFS++AIEAA+N+ +F LRE NTGTFPKGL++I +MA
Sbjct 1015 SSVVDKVEQIVLDCLEKHAREGFSEEAIEAAINSREFLLREFNTGTFPKGLAVIREMAAL 1074
Query 122 SNYDRDPVACLSFDASLKAVREKLKKEDEEGGYSLFQSLIRRFLLNNTHRVTVRLRADPE 181
DRDPV L F+ + +R +LK G +FQ+L+++F + N HR T+ LRADP+
Sbjct 1075 WTEDRDPVEGLRFEEHFEELRRRLK-----SGEPVFQNLLQKFFIGNPHRATIHLRADPD 1129
Query 182 MTAREAKQEKDNLSQIQKQLTPEVVESVLADQIALKQRQLTEDSEEALKTLPVLKLEDVA 241
AR QEK +S +Q L+ E ++ + LK RQ+ ED EAL TLP L LEDV
Sbjct 1130 EEARREAQEKAEISALQASLSSEKLDFLEKQTKELKARQMAEDPPEALATLPTLSLEDVD 1189
Query 242 DANNEVPYEVSALND 256
E+P + D
Sbjct 1190 KEGEEIPTSIEPFLD 1204
> ath:AT3G19170 ATPREP1 (PRESEQUENCE PROTEASE 1); metalloendopeptidase;
K06972
Length=1080
Score = 199 bits (506), Expect = 8e-51, Method: Compositional matrix adjust.
Identities = 106/257 (41%), Positives = 164/257 (63%), Gaps = 5/257 (1%)
Query 6 RMALQLLSYLLLGTSSSPLYKALSESGLGKSVIGGGLGLDLRHATFSVGLKGVAQEEDMV 65
++AL L +L+LGT +SPL K L ESGLG++++ GL +L F +GLKGV++E V
Sbjct 401 QLALGFLDHLMLGTPASPLRKILLESGLGEALVSSGLSDELLQPQFGIGLKGVSEEN--V 458
Query 66 EKVEKVIFRCLSSLSEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSIILDMATESNYD 125
+KVE++I L L+EEGF DA+EA++NT++F+LRE NTG+FP+GLS++L ++ YD
Sbjct 459 QKVEELIMDTLKKLAEEGFDNDAVEASMNTIEFSLRENNTGSFPRGLSLMLQSISKWIYD 518
Query 126 RDPVACLSFDASLKAVREKLKKEDEEGGYSLFQSLIRRFLLNNTHRVTVRLRADPEMTAR 185
DP L + LKA++ ++ EEG ++F LI + +LNN+HRVT+ ++ DPE +
Sbjct 519 MDPFEPLKYTEPLKALKTRIA---EEGSKAVFSPLIEKLILNNSHRVTIEMQPDPEKATQ 575
Query 186 EAKQEKDNLSQIQKQLTPEVVESVLADQIALKQRQLTEDSEEALKTLPVLKLEDVADANN 245
E +EK+ L +++ +T E + + LK +Q T D EAL+ +P L L D+
Sbjct 576 EEVEEKNILEKVKAAMTEEDLAELARATEELKLKQETPDPPEALRCVPSLNLGDIPKEPT 635
Query 246 EVPYEVSALNDVPLISH 262
VP EV +N V ++ H
Sbjct 636 YVPTEVGDINGVKVLRH 652
> ath:AT1G49630 ATPREP2 (ARABIDOPSIS THALIANA PRESEQUENCE PROTEASE
2); catalytic/ metal ion binding / metalloendopeptidase/
metallopeptidase/ zinc ion binding; K06972
Length=1080
Score = 182 bits (461), Expect = 1e-45, Method: Compositional matrix adjust.
Identities = 99/255 (38%), Positives = 160/255 (62%), Gaps = 5/255 (1%)
Query 6 RMALQLLSYLLLGTSSSPLYKALSESGLGKSVIGGGLGLDLRHATFSVGLKGVAQEEDMV 65
++AL L +L+LGT +SPL K L ESGLG++++ G+ +L FS+GLKGV+ +D V
Sbjct 400 QLALGFLDHLMLGTPASPLRKILLESGLGEALVNSGMEDELLQPQFSIGLKGVS--DDNV 457
Query 66 EKVEKVIFRCLSSLSEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSIILDMATESNYD 125
+KVE+++ L L++EGF DA+EA++NT++F+LRE NTG+ P+GLS++L + YD
Sbjct 458 QKVEELVMNTLRKLADEGFDTDAVEASMNTIEFSLRENNTGSSPRGLSLMLQSIAKWIYD 517
Query 126 RDPVACLSFDASLKAVREKLKKEDEEGGYSLFQSLIRRFLLNNTHRVTVRLRADPEMTAR 185
DP L ++ LK+++ ++ E+G S+F LI ++LNN H VT+ ++ DPE +
Sbjct 518 MDPFEPLKYEEPLKSLKARIA---EKGSKSVFSPLIEEYILNNPHCVTIEMQPDPEKASL 574
Query 186 EAKQEKDNLSQIQKQLTPEVVESVLADQIALKQRQLTEDSEEALKTLPVLKLEDVADANN 245
E +EK L +++ +T E + + L+ +Q T D +ALK +P L L D+
Sbjct 575 EEAEEKSILEKVKASMTEEDLTELARATEELRLKQETPDPPDALKCVPSLNLSDIPKEPI 634
Query 246 EVPYEVSALNDVPLI 260
VP EV +N V ++
Sbjct 635 YVPTEVGDINGVKVL 649
> tpv:TP02_0260 falcilysin
Length=1181
Score = 154 bits (390), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 91/259 (35%), Positives = 146/259 (56%), Gaps = 18/259 (6%)
Query 1 LSAADRMALQLLSYLLLGTSSSPLYKALSESGLGKSVIGGGLGLDLRHATFSVGLKGVAQ 60
+ D + Q+L YLLLGT S LYK L +SGLGK V+ G + + FS GLKGV
Sbjct 457 IDPVDNVGFQVLQYLLLGTPESVLYKGLIDSGLGKKVLVHGFLSGYKQSLFSFGLKGVDN 516
Query 61 -----EEDMVEKVEKVIFRCLSSLSEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSII 115
++++V+K E+V+F L + EEGF +DAI++ LN ++F +RELN+G++PKGL +I
Sbjct 517 TKFNSKDEIVKKFEEVVFGILRKIKEEGFKRDAIDSGLNLVEFEMRELNSGSYPKGLMLI 576
Query 116 LDMATESNYDRDPVACLSFDASLKAVREKLKKEDEEGGYSLFQSLIRRFLLNNTHRVTVR 175
+ ++ Y RDP A L FD+ +K +R ++ ++ + F +L+ + +LNN RVTV
Sbjct 577 DQIQSQLQYGRDPFALLRFDSLMKELRRRIFSDNPS---NYFINLMAKHILNNATRVTVH 633
Query 176 LRA------DPEMTAREAKQEKDNLSQIQKQLTPEVVESVLADQIALKQRQLTEDSEEAL 229
L A + E + AKQ + LS + K+ E+ E K + + D +
Sbjct 634 LHAVEASKYEKEFNKKIAKQLSERLSHLSKEQVDEMEEYYK----KFKNERESMDINDGS 689
Query 230 KTLPVLKLEDVADANNEVP 248
++L L+L D++ +P
Sbjct 690 ESLKTLELSDISREQETIP 708
> bbo:BBOV_II004890 18.m06407; peptidase M16 inactive domain containing
protein (EC:3.4.24.-); K06972
Length=1166
Score = 146 bits (368), Expect = 9e-35, Method: Compositional matrix adjust.
Identities = 84/250 (33%), Positives = 141/250 (56%), Gaps = 11/250 (4%)
Query 5 DRMALQLLSYLLLGTSSSPLYKALSESGLGKSVIGGGLGLDLRHATFSVGLKGVAQEE-- 62
D + +++L +LL+GTS S LYKAL +SGLGK V+G GL + + F +G+ G+ ++
Sbjct 451 DALGMEVLEHLLMGTSESYLYKALIKSGLGKKVVGSGLTNYFKQSNFIIGIAGIDPKQYD 510
Query 63 --DMVEKVEKVIFRCLSSLSEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSIILDMAT 120
+ + + ++ L + G K+AIEA++N ++F +RELNTGTFPKGL ++ M +
Sbjct 511 KANALATFDSIMNSTLLDMMNNGIKKEAIEASMNYIEFQIRELNTGTFPKGLMLVNLMQS 570
Query 121 ESNYDRDPVACLSFDASLKAVREKLKKEDEEGGYSLFQSLIRRFLLNNTHRVTVRLRA-D 179
+S Y +DP+ CL FD + +++++ + + FQ LI L+NN H+VTV ++A D
Sbjct 571 QSQYQKDPIECLYFDRFIAELKQRVANDSK-----YFQKLIDTHLVNNRHKVTVHMQAMD 625
Query 180 P-EMTAREAKQEKDNLSQIQKQLTPEVVESVLADQIALKQRQLTEDSEEALKTLPVLKLE 238
P E ++ + L LT V+++ + K D + L LP L L+
Sbjct 626 PKEFEKVTNERVRHELVASLSHLTKAQVDNMEQEYERFKAVCDNTDDRKTLDELPSLTLK 685
Query 239 DVADANNEVP 248
D+ + N +P
Sbjct 686 DINEKNELIP 695
> pfa:PF13_0322 fln; falcilysin; K06972
Length=1193
Score = 130 bits (326), Expect = 6e-30, Method: Composition-based stats.
Identities = 78/254 (30%), Positives = 129/254 (50%), Gaps = 13/254 (5%)
Query 5 DRMALQLLSYLLLGTSSSPLYKALSESGLGKSVIGGGLGLDLRHATFSVGLKGVAQEEDM 64
D L +++ LL+ T S LYKAL++ GLG +VI GL L FS+GLKG+ + +
Sbjct 412 DYFVLLIINNLLIHTPESVLYKALTDCGLGNNVIDRGLNDSLVQYIFSIGLKGIKRNNEK 471
Query 65 VE-------KVEKVIFRCLSSLSEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSIILD 117
++ +VE VI L + +EGF+K A+EA++N ++F L+E N T K + + +
Sbjct 472 IKNFDKVHYEVEDVIMNALKKVVKEGFNKSAVEASINNIEFILKEANLKT-SKSIDFVFE 530
Query 118 MATESNYDRDPVACLSFDASLKAVREKLKKEDEEGGYSLFQSLIRRFLLNNTHRVTVRLR 177
M ++ NY+RDP+ F+ L V+ K+K E + + + +NN HR + L
Sbjct 531 MTSKLNYNRDPLLIFEFEKYLNIVKNKIKNEP-----MYLEKFVEKHFINNAHRSVILLE 585
Query 178 ADPEMTAREAKQEKDNLSQIQKQLTPEVVESVLADQIALKQRQLTEDSEEALKTLPVLKL 237
D + EK L + + + E V+ + L + + E+S E L P++ +
Sbjct 586 GDENYAQEQENLEKQELKKRIENFNEQEKEQVIKNFEELSKYKNAEESPEHLNKFPIISI 645
Query 238 EDVADANNEVPYEV 251
D+ EVP V
Sbjct 646 SDLNKKTLEVPVNV 659
> hsa:10531 PITRM1, KIAA1104, MGC138192, MGC141929, MP1, PreP,
hMP1; pitrilysin metallopeptidase 1; K06972
Length=1037
Score = 95.9 bits (237), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 66/221 (29%), Positives = 113/221 (51%), Gaps = 14/221 (6%)
Query 21 SSPLYKALSESGLGKSVIGG-GLGLDLRHATFSVGLKGVAQEEDMVEKVEKVIFRCLSSL 79
+SP YKAL ESGLG G R A FSVGL+G+A+++ +E V +I R + +
Sbjct 358 NSPFYKALIESGLGTDFSPDVGYNGYTREAYFSVGLQGIAEKD--IETVRSLIDRTIDEV 415
Query 80 SEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSIILDMATESNYDRDPVACLSFDASLK 139
E+GF D IEA L+ ++ ++ +T GL + +A+ N+D DPV L L
Sbjct 416 VEKGFEDDRIEALLHKIEIQMKHQSTSF---GLMLTSYIASCWNHDGDPVELLKLGNQLA 472
Query 140 AVREKLKKEDEEGGYSLFQSLIRRFLLNNTHRVTVRLRADPEMTAREAKQEKDNLSQIQK 199
R+ L++ + Q ++++ NN H++T+ +R D + ++A+ E L Q +
Sbjct 473 KFRQCLQENPK-----FLQEKVKQYFKNNQHKLTLSMRPDDKYHEKQAQVEATKLKQKVE 527
Query 200 QLTPEVVESVLADQIALKQRQLTEDSEEALKTLPVLKLEDV 240
L+P + + + L+ +Q + LP LK+ D+
Sbjct 528 ALSPGDRQQIYEKGLELRSQQ---SKPQDASCLPALKVSDI 565
> mmu:69617 Pitrm1, 2310012C15Rik, AA410010, MP-1, MP1, Ntup1,
PreP, mKIAA1104; pitrilysin metallepetidase 1; K06972
Length=1036
Score = 92.4 bits (228), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 64/225 (28%), Positives = 115/225 (51%), Gaps = 14/225 (6%)
Query 17 LGTSSSPLYKALSESGLGKSVIGG-GLGLDLRHATFSVGLKGVAQEEDMVEKVEKVIFRC 75
+ +SP YKAL ESGLG G R A FSVGL+G+A+++ V+ V +++ R
Sbjct 354 IAGPNSPFYKALIESGLGTDFSPDVGYNGYTREAYFSVGLQGIAEKD--VKTVRELVDRT 411
Query 76 LSSLSEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSIILDMATESNYDRDPVACLSFD 135
+ + E+GF D IEA L+ ++ + + +F GL++ +A+ N+D DPV L
Sbjct 412 IEEVIEKGFEDDRIEALLHKIEIQTKH-QSASF--GLTLTSYIASCWNHDGDPVELLQIG 468
Query 136 ASLKAVREKLKKEDEEGGYSLFQSLIRRFLLNNTHRVTVRLRADPEMTAREAKQEKDNLS 195
+ L R+ LK+ + Q + ++ NN H++T+ ++ D + ++ + E + L
Sbjct 469 SQLTRFRKCLKENPK-----FLQEKVEQYFKNNQHKLTLSMKPDDKYYEKQTQMETEKLE 523
Query 196 QIQKQLTPEVVESVLADQIALKQRQLTEDSEEALKTLPVLKLEDV 240
Q L+P + + + L+ +Q + LP LK+ D+
Sbjct 524 QKVNSLSPADKQQIYEKGLELQTQQ---SKHQDASCLPALKVSDI 565
> dre:406808 pitrm1, wu:fa03d09, zgc:55469; pitrilysin metalloproteinase
1 (EC:3.4.24.-); K06972
Length=1023
Score = 88.6 bits (218), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 69/241 (28%), Positives = 120/241 (49%), Gaps = 16/241 (6%)
Query 21 SSPLYKALSESGLGKSVIGGGLGLD--LRHATFSVGLKGVAQEEDMVEKVEKVIFRCLSS 78
+SP YKAL E +G S G D R A+F++GL+G+A ED E V+ +I + +
Sbjct 353 NSPFYKALIEPKIG-SDFSSSAGFDGSTRQASFTIGLQGMA--EDDTETVKHIIAQTIDD 409
Query 79 LSEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSIILDMATESNYDRDPVACLSFDASL 138
+ GF ++ IEA L+ ++ ++ +T GL++ +A+ N+D DPV L S+
Sbjct 410 IIASGFEEEQIEALLHKIEIQMKHQSTSF---GLALASYIASLWNHDGDPVQLLKISESV 466
Query 139 KAVREKLKKEDEEGGYSLFQSLIRRFLLNNTHRVTVRLRADPEMTAREAKQEKDNLSQIQ 198
R+ LK+ Q ++ + NNTH++T+ + D ++A+ E+ L Q
Sbjct 467 SRFRQCLKENPR-----YLQEKVQHYFKNNTHQLTLSMSPDERFLEKQAEAEEQKLQQKI 521
Query 199 KQLTPEVVESVLADQIALKQRQLTEDSEEALKTLPVLKLEDVADANNEVPYEVSALNDVP 258
+ L+ E + + + L Q T +++A LP LK+ D+ P + A VP
Sbjct 522 QILSSEDRKDIYEKGLQLLAVQST--TQDA-SCLPALKVSDIEPIIPYTPVQPGAAGGVP 578
Query 259 L 259
+
Sbjct 579 V 579
> tpv:TP02_0257 falcilysin
Length=1119
Score = 83.2 bits (204), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 63/223 (28%), Positives = 110/223 (49%), Gaps = 19/223 (8%)
Query 1 LSAADRMALQLLSYLLLGTSSSPLYKALSESGLGKSVIGGGLGLDL---RHATFSVGLKG 57
L + D++AL++LSYLLL +S S L L S +G GL + +F G+ G
Sbjct 388 LDSVDKLALEVLSYLLLESSESVLLNKLVSSKFATRRVGPGLDEYFPAYEYLSFMFGVTG 447
Query 58 VAQEEDM----VEKVEKVIFRCLSSLSEEGFSKDAIEAALNTMQFT-------LRELNTG 106
V E + EK++ L+ + +GF++ A+EAALN ++F ++E G
Sbjct 448 VKYTEKTRDSNAKTFEKMVLEALTEVVTKGFNRKAVEAALNKVEFKHTEKKYEMKEHRRG 507
Query 107 TFPKGLSIILDMATESNYDRDPVACLSFDASLKAVREKLKKEDEEGGYSLFQSLIRRFLL 166
+P+GL+++ + +DP L F+ ++ ++ +D S +L+++ LL
Sbjct 508 YYPRGLALLRLVKPRYQEGKDPFELLRFEQLFPELKLRVFSDD---SCSYLSNLVKKHLL 564
Query 167 NNTHRVTVRLRA--DPEMTAREAKQEKDNLSQIQKQLTPEVVE 207
NN RVT+ L A + K+ D+L + +LT E V+
Sbjct 565 NNNTRVTLHLEAVESSKFEKEFNKKVSDHLRERVSKLTKEQVD 607
> xla:379672 pitrm1, MGC69133; pitrilysin metallopeptidase 1;
K06972
Length=1027
Score = 82.0 bits (201), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 68/254 (26%), Positives = 124/254 (48%), Gaps = 14/254 (5%)
Query 7 MALQLLSYLLLGTSSSPLYKALSESGLGKSV-IGGGLGLDLRHATFSVGLKGVAQEEDMV 65
L LLS L++ +SP YKAL E+ LG G R FS+GL+G+ +E+
Sbjct 338 FTLSLLSSLMVDGPNSPFYKALIEANLGTDFSPDTGFNNYTRETYFSIGLQGINKEDS-- 395
Query 66 EKVEKVIFRCLSSLSEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSIILDMATESNYD 125
EKV+ +I R ++ ++E+G + IEA L+ ++ ++ +T GL++ +A+ N++
Sbjct 396 EKVKHIINRTINEIAEQGIEPERIEALLHKLEIQMKHQSTSF---GLTLASYIASCWNHE 452
Query 126 RDPVACLSFDASLKAVREKLKKEDEEGGYSLFQSLIRRFLLNNTHRVTVRLRADPEMTAR 185
DPV L + R+ LK+ + Q ++++ N HR+ + + D + +
Sbjct 453 GDPVDLLKIGDKISRFRQCLKENPK-----FLQDKVKQYFQVNQHRMMLSMSPDEQHYDK 507
Query 186 EAKQEKDNLSQIQKQLTPEVVESVLADQIALKQRQLTEDSEEALKTLPVLKLEDVADANN 245
E + E++ L+Q K L+ E + + + L Q + LP LK+ D+
Sbjct 508 EEQLEEEKLTQKVKALSEEERKQIYEKGLELISLQ---SKPQDFSCLPALKVSDIEPQIP 564
Query 246 EVPYEVSALNDVPL 259
E++ DVP+
Sbjct 565 LTDLEIAYAGDVPV 578
> sce:YDR430C CYM1, MOP112; Cym1p (EC:3.4.24.-); K06972
Length=989
Score = 57.4 bits (137), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 58/247 (23%), Positives = 114/247 (46%), Gaps = 14/247 (5%)
Query 5 DRMALQLLSYLLLGTSSSPLYKALSESGLG-KSVIGGGLGLDLRHATFSVGLKGVAQEED 63
D L++L LL+ SS +Y+ L ESG+G + + G+ +VG++GV+ E
Sbjct 317 DTFLLKVLGNLLMDGHSSVMYQKLIESGIGLEFSVNSGVEPTTAVNLLTVGIQGVSDIEI 376
Query 64 MVEKVEKVIFRCLSSLSEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSIILDMATESN 123
+ V + L + E F + I+A + ++ + ++ GL ++ +
Sbjct 377 FKDTVNNIFQNLLET--EHPFDRKRIDAIIEQLELSKKDQKADF---GLQLLYSILPGWT 431
Query 124 YDRDPVACLSFDASLKAVREKLKKEDEEGGYSLFQSLIRRFLLNNTHRVTVRLRADPEMT 183
DP L F+ L+ R L E G +LFQ LIR+++++ T ++ E +
Sbjct 432 NKIDPFESLLFEDVLQRFRGDL----ETKGDTLFQDLIRKYIVHKP-CFTFSIQGSEEFS 486
Query 184 AREAKQEKDNLSQIQKQLTPEVVESVLADQIALKQRQLTEDSEEALKTLPVLKLEDVADA 243
+E+ L + L + +++ I L+++Q + +E L LP L+++D+ A
Sbjct 487 KSLDDEEQTRLREKITALDEQDKKNIFKRGILLQEKQ---NEKEDLSCLPTLQIKDIPRA 543
Query 244 NNEVPYE 250
++ E
Sbjct 544 GDKYSIE 550
> pfa:MAL8P1.139 conserved Plasmodium membrane protein, unknown
function
Length=5910
Score = 35.0 bits (79), Expect = 0.29, Method: Composition-based stats.
Identities = 21/75 (28%), Positives = 38/75 (50%), Gaps = 4/75 (5%)
Query 111 GLSIILDMATESNYDRDPVACLSFDASLKAVREKLKKEDEEG-GYSLFQSLIRRFLLNNT 169
GL ++ E +D P C+ + + ++E+ KKE+ E G S+F + FL +N
Sbjct 5704 GLQVVFIKEIEIKHDVSPKKCIC-EEKIHELKERRKKEEHENEGISIFN--LSYFLSSNY 5760
Query 170 HRVTVRLRADPEMTA 184
H+ R+D +M +
Sbjct 5761 HKKQSNTRSDEKMNS 5775
> pfa:MAL8P1.23 ubiquitin-protein ligase 1, putative; K10592 E3
ubiquitin-protein ligase HUWE1 [EC:6.3.2.19]
Length=8591
Score = 32.3 bits (72), Expect = 1.9, Method: Composition-based stats.
Identities = 31/110 (28%), Positives = 53/110 (48%), Gaps = 6/110 (5%)
Query 148 EDEEGGYSLFQSLIRRFL--LNNTHRVTVRLRADPEMTAREAKQEKDNLSQIQKQLTPEV 205
+++E +F S+ + +L L ++ R ++ R T R Q++ N+S I LTPE+
Sbjct 6509 DEKEQNIRIFNSIKKSYLEALPSSIRSEIKKRIIGNRTERTDVQDETNISFID-SLTPEL 6567
Query 206 VESVLADQIALKQRQLTEDSEEALKTLPVLKLED---VADANNEVPYEVS 252
VL+ R LT+D E + L ++ L + +ANN VS
Sbjct 6568 RRDVLSTASIRFIRTLTDDMIEEARNLRLVILNNRNSALNANNRNRQNVS 6617
> ath:AT1G72880 acid phosphatase survival protein SurE, putative;
K03787 5'-nucleotidase [EC:3.1.3.5]
Length=385
Score = 32.0 bits (71), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 27/47 (57%), Gaps = 0/47 (0%)
Query 80 SEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSIILDMATESNYDR 126
S+E KDA+ L + T+R++ G FPK S+ +++ T + ++
Sbjct 198 SQESHFKDAVGVCLPLINATIRDIAKGVFPKDCSLNIEIPTSPSSNK 244
> cel:F53F10.5 npp-11; Nuclear Pore complex Protein family member
(npp-11); K14306 nuclear pore complex protein Nup62
Length=805
Score = 31.6 bits (70), Expect = 3.3, Method: Composition-based stats.
Identities = 22/81 (27%), Positives = 39/81 (48%), Gaps = 8/81 (9%)
Query 67 KVEKVIFRCLSSLSEEGFSKDAIEAALNTMQFTLRELNTGT--------FPKGLSIILDM 118
++ +++ S + F D I +LNTMQ T+ + T T K L ++D+
Sbjct 718 QIAQMMLNVDSQMKCADFDLDQITKSLNTMQSTVLKTKTETPLEKTELIMKKQLQKLMDL 777
Query 119 ATESNYDRDPVACLSFDASLK 139
+T+ + RD + L D +LK
Sbjct 778 STQHDATRDKLNKLKDDHNLK 798
> hsa:9882 TBC1D4, AS160, DKFZp779C0666; TBC1 domain family, member
4
Length=1298
Score = 31.2 bits (69), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 33/122 (27%), Positives = 55/122 (45%), Gaps = 10/122 (8%)
Query 5 DRMALQLLSY---LLLGTSSSPLYKALSESGLGKSVIGGGLGLDLRHATFSVGLKGVAQE 61
D M+LQ+ Y LL LY L E+ + S+ L L + FS+G VA+
Sbjct 1047 DMMSLQIQMYQLSRLLHDYHRDLYNHLEENEISPSLYAAPWFLTLFASQFSLGF--VARV 1104
Query 62 EDMV-----EKVEKVIFRCLSSLSEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSIIL 116
D++ E + KV LSS ++ E + ++ TL ++NT K ++ +
Sbjct 1105 FDIIFLQGTEVIFKVALSLLSSQETLIMECESFENIVEFLKNTLPDMNTSEMEKIITQVF 1164
Query 117 DM 118
+M
Sbjct 1165 EM 1166
> tpv:TP04_0438 proteasome regulatory component; K03033 26S proteasome
regulatory subunit N3
Length=510
Score = 30.4 bits (67), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 33/131 (25%), Positives = 57/131 (43%), Gaps = 23/131 (17%)
Query 31 SGLGKSVIGGGL--------GLDLRHATFSVGLKGVAQEEDMVEK------VEKVI---F 73
S L +VI GGL ++L + +GLK V E+++ K +E VI
Sbjct 374 SRLRDNVIKGGLRKINLAYSKINLANVAHKLGLKSVEHTENIIAKAIHDGIIEAVIDHEN 433
Query 74 RCLSSLSEEGFSK--DAIEAALNTMQFTLR----ELNTGTFPKGLSIILDMATESNYDRD 127
+C++S K + + A +QF L+ + +P+ + + T SN D+D
Sbjct 434 QCVNSKVNVDLYKSYEPMRAFHKRIQFCLKLHSNAIQAMRYPEDPEVTKETKTTSNPDKD 493
Query 128 PVACLSFDASL 138
+ L +D L
Sbjct 494 QLESLDYDNEL 504
> ath:AT1G54110 cation exchanger, putative (CAX10)
Length=236
Score = 30.0 bits (66), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 27/105 (25%), Positives = 50/105 (47%), Gaps = 9/105 (8%)
Query 54 GLKGV--AQEEDMVEKVEKVIFRCL-----SSLSEEGFSKDAIEAALNTMQFTLRELNTG 106
GL V A+ + EK+E V+ R + + +S+E F+KD+ + ++ R N+
Sbjct 48 GLPRVTNAKVNEYYEKIEAVVSRIVAQVPHTEVSDEAFAKDSTNDSSPKVEDDTRTPNSP 107
Query 107 TFPKGLSIILDMATESNYDRDPVACLSFDASLKAVREKLKKEDEE 151
+ I+ + E +YD DP + D + +A K +K E+
Sbjct 108 QLRR--RIVPASSKEQSYDADPSKPIKLDTAAQAQVNKQRKLQED 150
Lambda K H
0.313 0.131 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9724477856
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40