bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
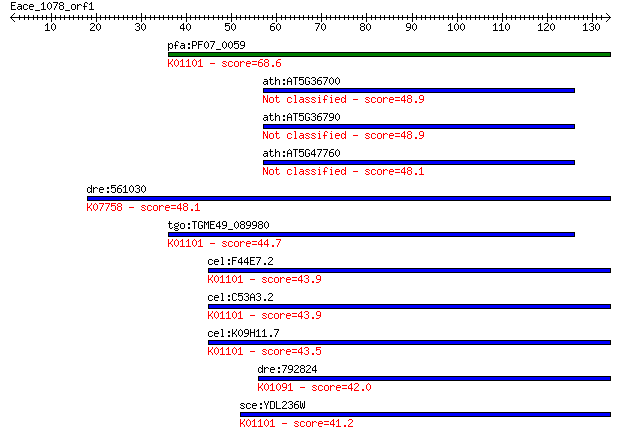
Query= Eace_1078_orf1
Length=133
Score E
Sequences producing significant alignments: (Bits) Value
pfa:PF07_0059 4-nitrophenylphosphatase, putative (EC:3.1.3.41)... 68.6 5e-12
ath:AT5G36700 PGLP1; PGLP1 (2-PHOSPHOGLYCOLATE PHOSPHATASE 1);... 48.9 4e-06
ath:AT5G36790 phosphoglycolate phosphatase, putative (EC:3.1.3... 48.9 4e-06
ath:AT5G47760 ATPGLP2 (ARABIDOPSIS THALIANA 2-PHOSPHOGLYCOLATE... 48.1 6e-06
dre:561030 zgc:194409 (EC:3.1.3.74); K07758 pyridoxal phosphat... 48.1 7e-06
tgo:TGME49_089980 4-nitrophenylphosphatase, putative (EC:3.1.3... 44.7 7e-05
cel:F44E7.2 hypothetical protein; K01101 4-nitrophenyl phospha... 43.9 1e-04
cel:C53A3.2 hypothetical protein; K01101 4-nitrophenyl phospha... 43.9 1e-04
cel:K09H11.7 hypothetical protein; K01101 4-nitrophenyl phosph... 43.5 2e-04
dre:792824 pgp, MGC56011, cb241, sb:cb241, wu:fb92c03, wu:fi40... 42.0 4e-04
sce:YDL236W PHO13; Alkaline phosphatase specific for p-nitroph... 41.2 8e-04
> pfa:PF07_0059 4-nitrophenylphosphatase, putative (EC:3.1.3.41);
K01101 4-nitrophenyl phosphatase [EC:3.1.3.41]
Length=322
Score = 68.6 bits (166), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 33/104 (31%), Positives = 60/104 (57%), Gaps = 6/104 (5%)
Query 36 QEQHLMCNGSSSI------QQQQQHKSRVYVVGEKGLLEELRSNGIDAFGGPEEAYEAID 89
+ +H++C + ++ + K ++YV+GEKG+ +EL ++ +D GG + + I
Sbjct 105 KREHIICTAYAVTKYLYDKEEYRLRKKKIYVIGEKGICDELDASNLDWLGGSNDNDKKII 164
Query 90 FARDPAVYVQPEVNYVVVGLDRDFYYKKLQTAQLYINDMHAKFI 133
D + V + VVVG+D + Y K+Q AQL IN+++A+FI
Sbjct 165 LKDDLEIIVDKNIGAVVVGIDFNINYYKIQYAQLCINELNAEFI 208
> ath:AT5G36700 PGLP1; PGLP1 (2-PHOSPHOGLYCOLATE PHOSPHATASE 1);
phosphoglycolate phosphatase (EC:3.1.3.18)
Length=362
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 27/69 (39%), Positives = 38/69 (55%), Gaps = 0/69 (0%)
Query 57 RVYVVGEKGLLEELRSNGIDAFGGPEEAYEAIDFARDPAVYVQPEVNYVVVGLDRDFYYK 116
+VYV+GE+G+L+EL G GGP++ I+ + +V VVVG DR F Y
Sbjct 165 KVYVIGEEGILKELELAGFQYLGGPDDGKRQIELKPGFLMEHDHDVGAVVVGFDRYFNYY 224
Query 117 KLQTAQLYI 125
K+Q L I
Sbjct 225 KIQYGTLCI 233
> ath:AT5G36790 phosphoglycolate phosphatase, putative (EC:3.1.3.18)
Length=362
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 27/69 (39%), Positives = 38/69 (55%), Gaps = 0/69 (0%)
Query 57 RVYVVGEKGLLEELRSNGIDAFGGPEEAYEAIDFARDPAVYVQPEVNYVVVGLDRDFYYK 116
+VYV+GE+G+L+EL G GGP++ I+ + +V VVVG DR F Y
Sbjct 165 KVYVIGEEGILKELELAGFQYLGGPDDGKRQIELKPGFLMEHDHDVGAVVVGFDRYFNYY 224
Query 117 KLQTAQLYI 125
K+Q L I
Sbjct 225 KIQYGTLCI 233
> ath:AT5G47760 ATPGLP2 (ARABIDOPSIS THALIANA 2-PHOSPHOGLYCOLATE
PHOSPHATASE 2); phosphoglycolate phosphatase/ protein serine/threonine
kinase (EC:3.1.3.18)
Length=301
Score = 48.1 bits (113), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 37/69 (53%), Gaps = 0/69 (0%)
Query 57 RVYVVGEKGLLEELRSNGIDAFGGPEEAYEAIDFARDPAVYVQPEVNYVVVGLDRDFYYK 116
+VYV+G +G+LEEL+ G GGPE+ + + + V VVVGLD + Y
Sbjct 105 KVYVIGGEGVLEELQIAGFTGLGGPEDGEKKAQWKSNSLFEHDKSVGAVVVGLDPNINYY 164
Query 117 KLQTAQLYI 125
KLQ L +
Sbjct 165 KLQYGTLCV 173
> dre:561030 zgc:194409 (EC:3.1.3.74); K07758 pyridoxal phosphatase
[EC:3.1.3.74]
Length=308
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 31/119 (26%), Positives = 55/119 (46%), Gaps = 7/119 (5%)
Query 18 PRQQQQQQQQQQKQQQLQQEQHLM---CNGSSSIQQQQQHKSRVYVVGEKGLLEELRSNG 74
PR+ Q+ + + +E+ C+ ++ ++ + + +VY +G G+L+ELR G
Sbjct 68 PRENYVQKFSRLGFADVAEEEIFSSAYCS-AAYLRDVARLQGKVYAIGGGGVLKELRDAG 126
Query 75 IDAFGGPEEAYEAIDFARDPAVYVQPEVNYVVVGLDRDFYYKKLQTAQLYINDMHAKFI 133
+ P E E P + P+V V+VG D F + KL A Y+ D F+
Sbjct 127 VPVVEEPAEQEEGTSIYNCP---LDPDVRAVLVGYDESFTFMKLAKACCYLRDAECLFL 182
> tgo:TGME49_089980 4-nitrophenylphosphatase, putative (EC:3.1.3.41
2.7.7.7); K01101 4-nitrophenyl phosphatase [EC:3.1.3.41]
Length=593
Score = 44.7 bits (104), Expect = 7e-05, Method: Composition-based stats.
Identities = 31/104 (29%), Positives = 46/104 (44%), Gaps = 14/104 (13%)
Query 36 QEQHLMCNGSSS---IQQQQQHKSRVYVVGEKGLLEELRSNGIDAFGGPEEAYEAIDFAR 92
E+ ++C ++ ++ H +V V+GE GL EE R G+ A E A +
Sbjct 342 HEEEMICTSYAAAEYMRLTHPHVKKVMVIGECGLKEEFREAGMVAVTAEEHASSPDAPSP 401
Query 93 DPAVY-----------VQPEVNYVVVGLDRDFYYKKLQTAQLYI 125
P++ + P V VVVG DR Y KL A LY+
Sbjct 402 APSISSERDFLDLTRALDPSVGAVVVGWDRQLSYVKLCLASLYL 445
> cel:F44E7.2 hypothetical protein; K01101 4-nitrophenyl phosphatase
[EC:3.1.3.41]
Length=335
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 50/94 (53%), Gaps = 7/94 (7%)
Query 45 SSSIQQQQQHKSRVYVVGEKGLLEELRSNGIDAFG-GPEEAYEAID----FARDPAVYVQ 99
+ ++ + RVY++GE+GL +E+ GI+ FG GPE+ + D F D + ++
Sbjct 122 ADTLHRAGLDGKRVYLIGEQGLRDEMDELGIEYFGHGPEKKQDEADGSGAFMYD--IKLE 179
Query 100 PEVNYVVVGLDRDFYYKKLQTAQLYINDMHAKFI 133
V VVVG ++ F Y K+ A Y+ + F+
Sbjct 180 ENVGAVVVGYEKHFDYIKMMKASNYLREEGVLFV 213
> cel:C53A3.2 hypothetical protein; K01101 4-nitrophenyl phosphatase
[EC:3.1.3.41]
Length=349
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 50/94 (53%), Gaps = 7/94 (7%)
Query 45 SSSIQQQQQHKSRVYVVGEKGLLEELRSNGIDAFG-GPEEAYEAID----FARDPAVYVQ 99
+ ++ + RVY++GE+GL +E+ GI+ FG GPE+ + D F D + ++
Sbjct 132 ADTLHRAGLDGKRVYLIGEQGLRDEMDELGIEYFGHGPEKKQDEADGSGAFMYD--IKLE 189
Query 100 PEVNYVVVGLDRDFYYKKLQTAQLYINDMHAKFI 133
V VVVG ++ F Y K+ A Y+ + F+
Sbjct 190 ENVGAVVVGYEKHFDYVKMMKASNYLREEGVLFV 223
> cel:K09H11.7 hypothetical protein; K01101 4-nitrophenyl phosphatase
[EC:3.1.3.41]
Length=322
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 50/94 (53%), Gaps = 7/94 (7%)
Query 45 SSSIQQQQQHKSRVYVVGEKGLLEELRSNGIDAFG-GPEEAYEAID----FARDPAVYVQ 99
+ ++ + RVY++GE+GL +E+ GI+ FG GPE+ + D F D + ++
Sbjct 105 ADTLHRAGLDGKRVYLIGEQGLRDEMDELGIEYFGHGPEKKQDEADGSGAFMYD--IKLE 162
Query 100 PEVNYVVVGLDRDFYYKKLQTAQLYINDMHAKFI 133
V VVVG ++ F Y K+ A Y+ + F+
Sbjct 163 ENVGAVVVGYEKHFDYVKMMKASNYLREEGVLFV 196
> dre:792824 pgp, MGC56011, cb241, sb:cb241, wu:fb92c03, wu:fi40b02,
zgc:56011; phosphoglycolate phosphatase (EC:3.1.3.18);
K01091 phosphoglycolate phosphatase [EC:3.1.3.18]
Length=306
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 41/80 (51%), Gaps = 5/80 (6%)
Query 56 SRVYVVGEKGLLEELRSNGIDAFG-GPEEAYEA-IDFARDPAVYVQPEVNYVVVGLDRDF 113
+VY++G K + +EL GI G GP+ ID+A P + EV V+VG D F
Sbjct 105 GKVYLIGSKAMKQELEEVGIQPVGVGPDLISGVQIDWANVP---LDQEVQAVLVGFDEHF 161
Query 114 YYKKLQTAQLYINDMHAKFI 133
Y KL A Y+ D +F+
Sbjct 162 SYMKLNRALQYLCDPDCQFV 181
> sce:YDL236W PHO13; Alkaline phosphatase specific for p-nitrophenyl
phosphate; also has protein phosphatase activity (EC:3.1.3.41);
K01101 4-nitrophenyl phosphatase [EC:3.1.3.41]
Length=312
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 25/85 (29%), Positives = 42/85 (49%), Gaps = 3/85 (3%)
Query 52 QQHKSRVYVVGEKGLLEELRSNGIDAFGGPEEAYEA-IDFARDPAVY--VQPEVNYVVVG 108
Q K +V+V GE G+ EEL+ G ++ GG + + D A+ P + + +V+ V+ G
Sbjct 107 QPGKDKVWVFGESGIGEELKLMGYESLGGADSRLDTPFDAAKSPFLVNGLDKDVSCVIAG 166
Query 109 LDRDFYYKKLQTAQLYINDMHAKFI 133
LD Y +L Y+ F+
Sbjct 167 LDTKVNYHRLAVTLQYLQKDSVHFV 191
Lambda K H
0.316 0.131 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2165519004
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40