bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1072_orf1
Length=199
Score E
Sequences producing significant alignments: (Bits) Value
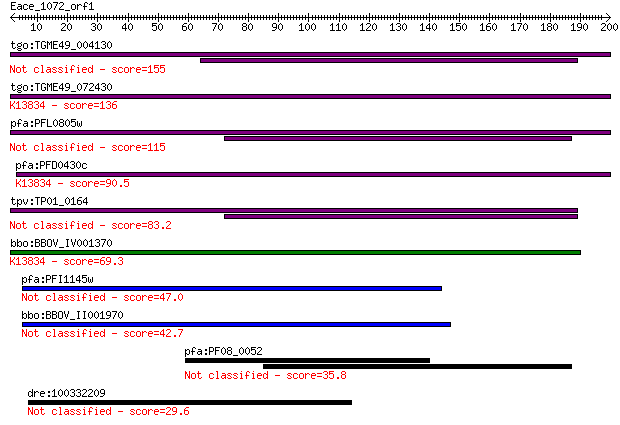
tgo:TGME49_004130 membrane-attack complex / perforin domain-co... 155 1e-37
tgo:TGME49_072430 membrane-attack complex / perforin domain-co... 136 5e-32
pfa:PFL0805w MAC/Perforin, putative 115 1e-25
pfa:PFD0430c MAC/Perforin, putative; K13834 sporozoite microne... 90.5 3e-18
tpv:TP01_0164 hypothetical protein 83.2 6e-16
bbo:BBOV_IV001370 21.m02755; MAC/perforin domain containing pr... 69.3 8e-12
pfa:PFI1145w MAC/Perforin, putative 47.0 4e-05
bbo:BBOV_II001970 18.m09950; mac/perforin domain containing me... 42.7 8e-04
pfa:PF08_0052 perforin like protein 5 35.8
dre:100332209 si:dkey-200m9.1-like 29.6 6.7
> tgo:TGME49_004130 membrane-attack complex / perforin domain-containing
protein
Length=1054
Score = 155 bits (391), Expect = 1e-37, Method: Composition-based stats.
Identities = 82/206 (39%), Positives = 121/206 (58%), Gaps = 9/206 (4%)
Query 1 GFAMCAPHERVLLGFALELNFLDENAIAARAGIKACAPGREKCVGLERPAKEGDDARIFV 60
GFA C + V+LGFA+ LNF + R I +C PGREKC G+ + E D+ RI++
Sbjct 734 GFAKCPEGQVVILGFAMHLNFKEPGTDNFR--IISCPPGREKCDGVGTASSETDEGRIYI 791
Query 61 LCGPEPVAGLEQVVVQSPLKA-----VAVCPQGSMILTGFALSLTGGREGPKKTAFFPCR 115
LCG EP+ ++QVV +SP A A CP ++++ GF +S+ GG +G + C
Sbjct 792 LCGEEPINEIQQVVAESPAHAGASVLEASCPDETVVVGGFGISVRGGSDGLDSFSIESCT 851
Query 116 AGLPSCTALGLRGTQQNMVWIVCVDEATPGLQTVTNVGEVVT-GVATKHAY-TDGIVKAS 173
G CT RG+++N +W++CVD+ PGL+ + NV E+ + G A K A +DG V
Sbjct 852 TGQTICTKAPTRGSEKNFLWMMCVDKQYPGLRELVNVAELGSHGNANKRAVNSDGNVDVK 911
Query 174 CPPNLGVALGFGLELHTRMPRVREKF 199
CP N + LG+ +E HT + VR+KF
Sbjct 912 CPANSSIVLGYVMEAHTNLQFVRDKF 937
Score = 50.1 bits (118), Expect = 5e-06, Method: Composition-based stats.
Identities = 32/127 (25%), Positives = 52/127 (40%), Gaps = 10/127 (7%)
Query 64 PEPVAGLEQVVVQSPLKAVAVCPQGSMILTGFALSLTGGREGPKKTAFFPCRAGLPSCTA 123
P+ + QV P A CP+G +++ GFA+ L G C G C
Sbjct 717 PKQLTQATQVAWSGPPPGFAKCPEGQVVILGFAMHLNFKEPGTDNFRIISCPPGREKCDG 776
Query 124 LGLRG--TQQNMVWIVCVDEATPGLQTVTNVGEVVTGVATKHAYTDGIVKASCPPNLGVA 181
+G T + ++I+C +E +Q V G + +++ASCP V
Sbjct 777 VGTASSETDEGRIYILCGEEPINEIQQVVAESPAHAGAS--------VLEASCPDETVVV 828
Query 182 LGFGLEL 188
GFG+ +
Sbjct 829 GGFGISV 835
> tgo:TGME49_072430 membrane-attack complex / perforin domain-containing
protein ; K13834 sporozoite microneme protein 2
Length=854
Score = 136 bits (342), Expect = 5e-32, Method: Composition-based stats.
Identities = 81/207 (39%), Positives = 110/207 (53%), Gaps = 10/207 (4%)
Query 1 GFAMCAPHERVLLGFALELNF---LDENAIAARAGIKACAPGREKCVGLERPAKEGDDAR 57
G+A C + VL GFA+ NF + N A I C GREKC G+ GDD R
Sbjct 597 GYARCPREQVVLFGFAMRFNFKVTISNNL--ANYHIAPCTAGREKCDGIGAEEAAGDDER 654
Query 58 IFVLCGPEPVAGLEQVVVQSPLK---AVAVCPQGSMILTGFALSLTGGREGPKKTAFFPC 114
I++ CGPE V QVV ++ AVA CP+ ++I GF +S+ G + T PC
Sbjct 655 IYMACGPEVVNEFYQVVAETEAGENVAVATCPEDTVIAFGFGISIGTGFYSSENTQVEPC 714
Query 115 RAGLPSCTALGLRGTQQNMVWIVCVDEATPGLQTVTNVGEVVT-GVA-TKHAYTDGIVKA 172
AG CT T ++ VW+VC +++ PG+ + N+ EV T G A ++ TDGIV
Sbjct 715 TAGQTRCTKARTSNTVKSYVWMVCAEKSFPGIAQLNNIAEVGTRGKANSRMKNTDGIVNV 774
Query 173 SCPPNLGVALGFGLELHTRMPRVREKF 199
SC + LG LE+HT MP VR+ F
Sbjct 775 SCGADERTLLGLALEVHTHMPSVRKAF 801
> pfa:PFL0805w MAC/Perforin, putative
Length=1073
Score = 115 bits (287), Expect = 1e-25, Method: Composition-based stats.
Identities = 72/207 (34%), Positives = 98/207 (47%), Gaps = 13/207 (6%)
Query 1 GFAMCAPHERVLLGFALELNFLDENAIAARAGIKACAPGREKCVGLERPAKEGDDARIFV 60
G C +L+GF++ LNF +++ GI C P +E C G K D RIF
Sbjct 825 GLLTCPIGTTILMGFSINLNFYKNKYLSSTNGITLCEPMKESCSG-NGFEKNYSDIRIFA 883
Query 61 LCGPEPVAGLEQVVVQSPL-KAVAVCPQGSMILTGFALSLTGGREGPKKTAFFPCRAGLP 119
LC +P + QVV Q K A CP +IL GFAL G K +PCR G
Sbjct 884 LCTNKPFDFITQVVQQGEAPKISASCPGELVILFGFALMKGIGSSSANKIDIYPCRTGQN 943
Query 120 SCTA-LGLRGTQQNMVWIVCVDEATPGLQ------TVTNVGEVVTGVATKHAYTDGIVKA 172
SC A L +Q+M+++ CVD+ T GL+ N+G+V+ + +DG +
Sbjct 944 SCEAVLQNHKFKQSMIYLACVDKTTNGLEYLQTYSKTKNLGDVI----SDKYKSDGYLNF 999
Query 173 SCPPNLGVALGFGLELHTRMPRVREKF 199
SCP N + GF LE HT R F
Sbjct 1000 SCPQNNTLVFGFSLEFHTNFQATRNNF 1026
Score = 33.1 bits (74), Expect = 0.68, Method: Composition-based stats.
Identities = 28/118 (23%), Positives = 46/118 (38%), Gaps = 15/118 (12%)
Query 72 QVVVQSPLKAVAVCPQGSMILTGFALSLT--GGREGPKKTAFFPCRAGLPSCTALGLRGT 129
Q + P + CP G+ IL GF+++L + C SC+ G
Sbjct 816 QKLYAGPPPGLLTCPIGTTILMGFSINLNFYKNKYLSSTNGITLCEPMKESCSGNGFEKN 875
Query 130 QQNM-VWIVCVDEATPGLQTVTNVGEVVTGVATKHAYTDGIVKASCPPNLGVALGFGL 186
++ ++ +C ++ + V GE A K + ASCP L + GF L
Sbjct 876 YSDIRIFALCTNKPFDFITQVVQQGE-----APK-------ISASCPGELVILFGFAL 921
> pfa:PFD0430c MAC/Perforin, putative; K13834 sporozoite microneme
protein 2
Length=842
Score = 90.5 bits (223), Expect = 3e-18, Method: Composition-based stats.
Identities = 59/204 (28%), Positives = 94/204 (46%), Gaps = 9/204 (4%)
Query 3 AMCAPHERVLLGFALELNFLDENAIAARAGIKACAPGREKCVGLERPAKEGDDARIFVLC 62
A C + V+ GF+L+ NF D I+ C G C + + + D + +++ C
Sbjct 604 AQCPHGKVVMFGFSLKQNFWDNTNALKGYNIEVCEAGSNSCTSKQGSSNKYDTSYLYMEC 663
Query 63 GPEPVAGLEQVVVQSPLKAVAV-CPQGSMILTGFALSLTGGREGPKKTAF-FPCRAGLPS 120
G +P+ EQV+ +S V CP IL GF +S + GR + + PC G+ S
Sbjct 664 GDQPLPFSEQVISESTSTYNTVKCPNDYSILLGFGISSSSGRINSAEYVYSTPCIPGMKS 723
Query 121 CTALGLRGTQQNMVWIVCVDEATPGLQTVTNVGEVVTG-----VATKHAYTDGIVKASCP 175
C+ Q++ ++++CVD T V N+ V V Y+DG + +CP
Sbjct 724 CSLNMNNDNQKSYIYVLCVD--TTIWSGVNNLSLVALDGAHGKVNRSKKYSDGELVGTCP 781
Query 176 PNLGVALGFGLELHTRMPRVREKF 199
+ V GF +E HT P V+ F
Sbjct 782 LDGTVLTGFKVEFHTSSPYVQTPF 805
> tpv:TP01_0164 hypothetical protein
Length=1182
Score = 83.2 bits (204), Expect = 6e-16, Method: Composition-based stats.
Identities = 64/204 (31%), Positives = 91/204 (44%), Gaps = 31/204 (15%)
Query 1 GFAMCAPHERVLLGFALEL----NFLDENAIAARAGIKACAPGREKC-VGLERPAKEGDD 55
G A+C +++GF+L + N + +N I C G EKC V + P E
Sbjct 944 GSAICPNKTVIIMGFSLSILKKKNIVGKNEFTLH--ITQCPVGEEKCIVSSDNPMSE--- 998
Query 56 ARIFVLCGPEPVAGLEQVV---VQSPLKAVAVCPQGSMILTGFALSLTGGREGPKKTAFF 112
+RI+ +CG + + L Q + P A A CP G I GFALS+ G T +
Sbjct 999 SRIWAVCGEDTIPLLNQQTSSEIDEP--ATATCPVGYSIAYGFALSVPKGNVA-LNTDSY 1055
Query 113 PCRAGLPSCTALGLRGTQQNMVWIVCVDEATPGLQTVTNVGEVVTGVATKH--------A 164
CR+G SCT T N VWI CV+ P L ++N V +++H A
Sbjct 1056 ACRSGTQSCTHESTDKTATNAVWIACVENGAPQLSEISN---HVVSTSSRHCSKKTKDNA 1112
Query 165 YTDGIVKASCPPNLGVALGFGLEL 188
Y D SCP N + G+ L+
Sbjct 1113 YDDN----SCPINSKLIAGWSLDF 1132
Score = 44.7 bits (104), Expect = 2e-04, Method: Composition-based stats.
Identities = 31/123 (25%), Positives = 49/123 (39%), Gaps = 20/123 (16%)
Query 72 QVVVQSPLKAVAVCPQGSMILTGFALSLTGGREGPKKTAF----FPCRAGLPSCTALGLR 127
Q+V A+CP ++I+ GF+LS+ + K F C G C
Sbjct 935 QIVYSGNKSGSAICPNKTVIIMGFSLSILKKKNIVGKNEFTLHITQCPVGEEKCIVSSDN 994
Query 128 GTQQNMVWIVCVDEATPGL--QTVTNVGEVVTGVATKHAYTDGIVKASCPPNLGVALGFG 185
++ +W VC ++ P L QT + + E T A+CP +A GF
Sbjct 995 PMSESRIWAVCGEDTIPLLNQQTSSEIDEPAT--------------ATCPVGYSIAYGFA 1040
Query 186 LEL 188
L +
Sbjct 1041 LSV 1043
> bbo:BBOV_IV001370 21.m02755; MAC/perforin domain containing
protein; K13834 sporozoite microneme protein 2
Length=978
Score = 69.3 bits (168), Expect = 8e-12, Method: Composition-based stats.
Identities = 55/195 (28%), Positives = 87/195 (44%), Gaps = 15/195 (7%)
Query 1 GFAMCAPHERVLLGFALELNFLDENAIAA---RAGIKACAPGREKCVGLERPAKEGDDAR 57
G A+C + +++GF++ ++ + + C G+EKC+ P EG R
Sbjct 685 GSAVCPNGKVIMMGFSVVISSSRATVFSKPQYTISMTPCPIGQEKCMVSVPPGAEG---R 741
Query 58 IFVLCGPEPVAGL-EQVVVQSPLKAVAVCPQGSMILTGFALSLTGGREGPKKTAF--FPC 114
+++LCG E + L ++ V + A A CP I GF LS+ +G K T + C
Sbjct 742 VWILCGSESIPLLIQETNVANNEAATAQCPDEYAIAFGFGLSIP---DGAKLTPVDCYAC 798
Query 115 RAGLPSCTALGLRGTQQNMVWIVCVDEATPGLQTVTNVGEVVTGVATKHAYTDGIVKASC 174
RAG SCT + + N VWI CV++ P L ++ N V C
Sbjct 799 RAGQQSCTQASPK-SPYNAVWIACVEKNAPELGSIVNQARVSLSEGCSSKLPQTTADNIC 857
Query 175 PPNLGVALGFGLELH 189
P +G L G E++
Sbjct 858 P--VGTLLIAGWEMN 870
> pfa:PFI1145w MAC/Perforin, putative
Length=821
Score = 47.0 bits (110), Expect = 4e-05, Method: Composition-based stats.
Identities = 40/144 (27%), Positives = 65/144 (45%), Gaps = 15/144 (10%)
Query 5 CAPHERVLLGFALELNFLDENAIAARAGIKACAPGREKCVGLERPAKEGDDAR--IFVLC 62
C + LLGF+L + ++ + + +C +KC +K D+A IF +C
Sbjct 605 CEEKQNFLLGFSLSIP--NDLSNLKDFYLNSCDEDSDKCY-----SKMSDNAYSYIFAMC 657
Query 63 GPEPVAGLEQVVVQS-PLKAVAVCPQGSMILTGFALSLTGGREGPKKTAFFPCRAGLPSC 121
E + EQ V L + + +IL GF +S+ + P + +PC+ G SC
Sbjct 658 KEEMIPFFEQKVKSGVGLLTLECSEKNQVILFGFGISVLNTND-PISISLYPCKYGKASC 716
Query 122 TALGLRGTQQNMV--WIVCVDEAT 143
+ G T Q+ V WIVC E +
Sbjct 717 SMQG--STDQSAVGLWIVCAHEES 738
> bbo:BBOV_II001970 18.m09950; mac/perforin domain containing
membrane protein
Length=559
Score = 42.7 bits (99), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 38/146 (26%), Positives = 58/146 (39%), Gaps = 17/146 (11%)
Query 5 CAPHERVLLGFALELNFLDENAIAARA-GIKACAPGREKCVGLERPAKEGDDARIFVLCG 63
C +++V+ GF LE+ I R + C C ER K D I++LCG
Sbjct 399 CPHNDKVIFGFILEM------EIEQRTFSVYQCPTDAYSC-SKERQKKC--DIVIWMLCG 449
Query 64 PEPVAGLEQVVVQ---SPLKAVAVCPQGSMILTGFALSLTGGREGPKKTAFFPCRAGLPS 120
+ Q S + C G +LTGF ++ + + PC G
Sbjct 450 SSMGLNVMQYAHNFDASNSEKEVKCLSGYKLLTGF-IAESSPEKDKSLANLIPCHTGADL 508
Query 121 CTALGLRGTQQNMVWIVCVDEATPGL 146
C + + + +W VC+DE PGL
Sbjct 509 CRS---NSSLETHIWAVCIDERLPGL 531
> pfa:PF08_0052 perforin like protein 5
Length=676
Score = 35.8 bits (81), Expect = 0.11, Method: Composition-based stats.
Identities = 21/83 (25%), Positives = 34/83 (40%), Gaps = 4/83 (4%)
Query 59 FVLCGPEPVAGLEQVVVQSPLKA--VAVCPQGSMILTGFALSLTGGREGPKKTAFFPCRA 116
++LC E + + Q++ ++ + A CP I GF SLT + PC +
Sbjct 505 WILCSKENRSEIHQILTKNTFQGNGKASCPYNMKI--GFGFSLTFQKSINTNIKIEPCES 562
Query 117 GLPSCTALGLRGTQQNMVWIVCV 139
C L + Q WI C+
Sbjct 563 NKKECKRTNLASSSQTYFWINCL 585
Score = 30.0 bits (66), Expect = 5.6, Method: Composition-based stats.
Identities = 26/103 (25%), Positives = 41/103 (39%), Gaps = 13/103 (12%)
Query 85 CPQGSMILTGFALSLTGGREGPKKTAFFPCRAGLPSCTALGLRGTQQ-NMVWIVCVDEAT 143
C G IL+GF L+ + C + + + + + WI+C E
Sbjct 455 CKNGDKILSGFILT-NKKKSYEDNHIIHMCPSNTVCSSGINIESDKNFEFSWILCSKE-- 511
Query 144 PGLQTVTNVGEVVTGVATKHAYTDGIVKASCPPNLGVALGFGL 186
N E+ + TK+ + G KASCP N+ + GF L
Sbjct 512 -------NRSEI-HQILTKNTF-QGNGKASCPYNMKIGFGFSL 545
> dre:100332209 si:dkey-200m9.1-like
Length=254
Score = 29.6 bits (65), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 34/109 (31%), Positives = 46/109 (42%), Gaps = 17/109 (15%)
Query 7 PHERVLLGFA-LELNFL-DENAIAARAGIKACAPGREKCVGLERPAKEGDDARIFVLCGP 64
PH + +G A L LN + D+ I A G AC+ + +GL P E + I L
Sbjct 130 PHFSICIGIAALLLNDIKDQMFIMASEGQLACS---DNSLGLLSPYME--QSSIMDL--- 181
Query 65 EPVAGLEQVVVQSPLKAVAVCPQGSMILTGFALSLTGGREGPKKTAFFP 113
E G E + AV +CPQG A + G E + FFP
Sbjct 182 EATRGFEAIPQVKCTNAVNLCPQG-------ATTCMGSAEAQFQCRFFP 223
Lambda K H
0.322 0.139 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5931269072
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40