bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1016_orf2
Length=239
Score E
Sequences producing significant alignments: (Bits) Value
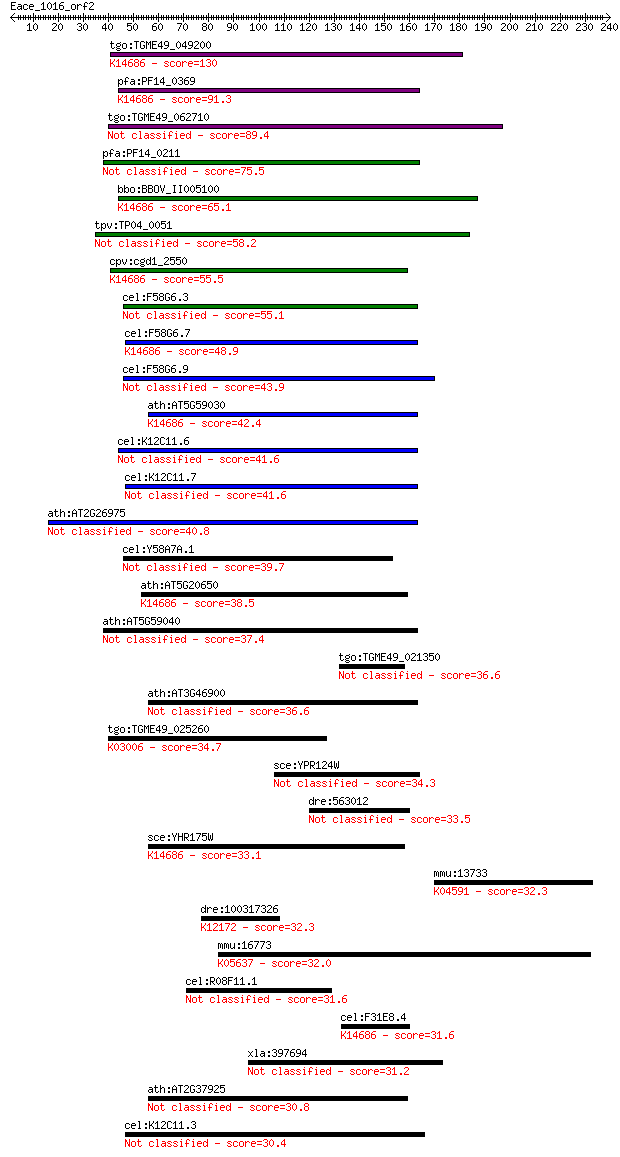
tgo:TGME49_049200 ctr copper transporter domain-containing pro... 130 3e-30
pfa:PF14_0369 copper transporter putative; K14686 solute carri... 91.3 3e-18
tgo:TGME49_062710 ctr copper transporter domain-containing pro... 89.4 1e-17
pfa:PF14_0211 Ctr copper transporter domain containing protein... 75.5 1e-13
bbo:BBOV_II005100 18.m09998; hypothetical protein; K14686 solu... 65.1 2e-10
tpv:TP04_0051 polymorphic immunodominant molecule 58.2 3e-08
cpv:cgd1_2550 copper transporter, 3 transmembrane domain, cons... 55.5 2e-07
cel:F58G6.3 hypothetical protein 55.1 2e-07
cel:F58G6.7 hypothetical protein; K14686 solute carrier family... 48.9 1e-05
cel:F58G6.9 hypothetical protein 43.9 5e-04
ath:AT5G59030 COPT1; COPT1 (copper transporter 1); copper ion ... 42.4 0.002
cel:K12C11.6 hypothetical protein 41.6 0.002
cel:K12C11.7 hypothetical protein 41.6 0.002
ath:AT2G26975 copper transporter, putative 40.8 0.005
cel:Y58A7A.1 hypothetical protein 39.7 0.009
ath:AT5G20650 COPT5; COPT5; copper ion transmembrane transport... 38.5 0.021
ath:AT5G59040 COPT3; COPT3; copper ion transmembrane transport... 37.4 0.051
tgo:TGME49_021350 hypothetical protein 36.6 0.091
ath:AT3G46900 COPT2; COPT2; copper ion transmembrane transport... 36.6 0.092
tgo:TGME49_025260 DNA-directed RNA polymerase II largest subun... 34.7 0.35
sce:YPR124W CTR1; Ctr1p 34.3 0.40
dre:563012 sb:cb797; si:ch211-282j22.4; K14687 solute carrier ... 33.5 0.62
sce:YHR175W CTR2; Ctr2p; K14686 solute carrier family 31 (copp... 33.1 0.86
mmu:13733 Emr1, DD7A5-7, EGF-TM7, F4/80, Gpf480, Ly71, TM7LN3;... 32.3 1.5
dre:100317326 si:ch1073-55a19.2; K12172 E3 SUMO-protein ligase... 32.3 1.7
mmu:16773 Lama2, 5830440B04, KIAA4087, dy, mKIAA4087, mer, mer... 32.0 2.0
cel:R08F11.1 hypothetical protein 31.6 2.7
cel:F31E8.4 hypothetical protein; K14686 solute carrier family... 31.6 2.9
xla:397694 slc31a2, copt2, ctr2, xem1; solute carrier family 3... 31.2 3.5
ath:AT2G37925 COPT4; COPT4; copper ion transmembrane transport... 30.8 4.0
cel:K12C11.3 hypothetical protein 30.4 5.9
> tgo:TGME49_049200 ctr copper transporter domain-containing protein
; K14686 solute carrier family 31 (copper transporter),
member 1
Length=235
Score = 130 bits (328), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 67/141 (47%), Positives = 89/141 (63%), Gaps = 1/141 (0%)
Query 41 GLPLPMWFEASSEVLLLFSWWNARTTAQYVVCCLCCIAFGFVSIALKVLRRVSEVRLKMA 100
G+PLPM FE S+ V+ LF W TT Q+ C+ GF+ + LKV+RR E L
Sbjct 71 GMPLPMAFEVSTRVIYLFEDWPTETTTQFAGACVATCILGFICVILKVVRRYVEKSLVSQ 130
Query 101 EKKHKPTLMLGSLPVFHNALRASVAFLNYSWDYMLMLVAMTFNVGIFLSMLGGMALGFLT 160
E K L+ GS P++ N++R VAF+NYSWDYMLML++MTFNVGIFLS+L G+ALGFL
Sbjct 131 ENVGKTKLIFGSFPLYSNSVRFLVAFVNYSWDYMLMLLSMTFNVGIFLSLLLGIALGFLF 190
Query 161 IGRYLGFSL-QPKNIVSGCEC 180
+G + + K C+C
Sbjct 191 LGDLMSVEVGSTKKPWKPCQC 211
> pfa:PF14_0369 copper transporter putative; K14686 solute carrier
family 31 (copper transporter), member 1
Length=235
Score = 91.3 bits (225), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 42/121 (34%), Positives = 76/121 (62%), Gaps = 5/121 (4%)
Query 44 LPMWFEASSEVLLLFSWWNARTTAQYVVCCLCCIAFGFVSIALKVLRRVSEVRLKMAEKK 103
+PM F+ ++ ++LF+ W ++ Y + + C FG +S+ KV+R + ++ A K
Sbjct 91 MPMSFQLTTHTIILFNKWETKSALSYYISLVLCFFFGIISVGFKVVR----LNVEQALPK 146
Query 104 HKPTLMLGSLPVF-HNALRASVAFLNYSWDYMLMLVAMTFNVGIFLSMLGGMALGFLTIG 162
+ T + SL +F +N+ R ++F+ YSWDY+LML+ MTFNVG+F++++ G++ GF G
Sbjct 147 TEDTNIFKSLVLFKNNSYRMLLSFVIYSWDYLLMLIVMTFNVGLFVAVVLGLSFGFFIFG 206
Query 163 R 163
Sbjct 207 N 207
> tgo:TGME49_062710 ctr copper transporter domain-containing protein
Length=375
Score = 89.4 bits (220), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 57/181 (31%), Positives = 88/181 (48%), Gaps = 34/181 (18%)
Query 40 CGLPLPMWFEASSEVLLLFSWWNARTTAQYVVCCLCCIAFGFVSIALKVLR--------- 90
CG+ +PM F+ S ++LF W QYV+ L C+ G +S+ LKVLR
Sbjct 203 CGV-MPMSFQNSLHTVILFHSWETLERWQYVLSLLTCVVLGMLSVVLKVLRLRLEFFLAK 261
Query 91 --RVSEVRLKMAEKKHKP-------------TLMLGSLPVFHNALRASVAFLNYSWDYML 135
R +E ++ + K K + G+ P+ N+ R AF+ Y +DY+L
Sbjct 262 RDRAAEDAQRVEKLKEKEGQSSAASPSSAIVERLCGNFPLKQNSWRMLEAFVIYGYDYLL 321
Query 136 MLVAMTFNVGIFLSMLGGMALGFLTIGRYLGFSLQPKNIVSGCECDEDFSCGCHKGQSCT 195
ML+ MT+NVG+F ++ GG+ALGF G L +Q + + E D ++G C
Sbjct 322 MLIVMTYNVGLFFAVTGGLALGFFCFGHLL--RIQAEKEENSLEED-------YRGDPCC 372
Query 196 C 196
C
Sbjct 373 C 373
> pfa:PF14_0211 Ctr copper transporter domain containing protein,
putative
Length=160
Score = 75.5 bits (184), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 41/127 (32%), Positives = 70/127 (55%), Gaps = 4/127 (3%)
Query 38 NKCGLPLPMWFEASSEVLLLFSWWNARTTAQYVVCCLCCIAFGFVSIALKVL-RRVSEVR 96
N G+ LPM+F + + +LF + + Q++ C + CI GF S+ +KVL +R+
Sbjct 35 NDDGVMLPMYFSNNENIKMLFDIFQVKNRYQFIFCNILCIIMGFFSVYIKVLKKRLHHNV 94
Query 97 LKMAEKKHKPTLMLGSLPVFHNALRASVAFLNYSWDYMLMLVAMTFNVGIFLSMLGGMAL 156
K+A+ + + N ++FL+Y+ DY+LML+ MTFN IFLS++ G++
Sbjct 95 QKVADGGDGSYVNMSPC---QNVNYGFLSFLHYTIDYLLMLIVMTFNPYIFLSIMTGLSS 151
Query 157 GFLTIGR 163
+L G
Sbjct 152 AYLFYGH 158
> bbo:BBOV_II005100 18.m09998; hypothetical protein; K14686 solute
carrier family 31 (copper transporter), member 1
Length=297
Score = 65.1 bits (157), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 42/143 (29%), Positives = 63/143 (44%), Gaps = 8/143 (5%)
Query 44 LPMWFEASSEVLLLFSWWNARTTAQYVVCCLCCIAFGFVSIALKVLRRVSEVRLKMAEKK 103
+PM+FE + + ++LF +W T QY V +++ LK R L
Sbjct 163 MPMYFENTVKTVILFHFWKTTTGTQYAVSLFFIFVLSLMTVFLKAFRNKLNCALLQRPNG 222
Query 104 HKPTLMLGSLPVFHNALRASVAFLNYSWDYMLMLVAMTFNVGIFLSMLGGMALGFLTIGR 163
+ PT+ G + + +AF+ D+ +MLV MTFNVGI L + ALG++
Sbjct 223 YHPTVKYGIMYI--------LAFVVTFMDFAMMLVVMTFNVGIVLVVCSAYALGYIFTCC 274
Query 164 YLGFSLQPKNIVSGCECDEDFSC 186
LG S EC D C
Sbjct 275 PLGLSEAANGSRCTQECPADCCC 297
> tpv:TP04_0051 polymorphic immunodominant molecule
Length=480
Score = 58.2 bits (139), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 43/150 (28%), Positives = 68/150 (45%), Gaps = 14/150 (9%)
Query 35 TMLNKCGLPLPMWFEASSEVLLLFSWWNARTTAQYVVCCLCCIAFGFVSIALKVLRRVSE 94
T + CG +F + +V ++F WW QY + L F +S LK R V
Sbjct 342 TPFHGCG----QFFTNTHKVTVIFHWWLCEKPWQYALTLLTLFGFALLSPCLKAYREV-- 395
Query 95 VRLKMAEKKHKPTLMLGSLPVFHNALRASVAFLNYSWDYMLMLVAMTFNVGIFLSMLGGM 154
+R K + + L + +A Y+ D++LMLV MTFNVG+F +++ G
Sbjct 396 LRAKAVR-----SFIFDCL--LTHLFLFLIALCAYALDFLLMLVVMTFNVGVFFAVILGY 448
Query 155 ALGF-LTIGRYLGFSLQPKNIVSGCECDED 183
++G+ L+ Y QP S +ED
Sbjct 449 SVGYVLSSLAYSTLRTQPNRSNSFSRINED 478
> cpv:cgd1_2550 copper transporter, 3 transmembrane domain, conserved
in metazoa and apiacomplexa ; K14686 solute carrier
family 31 (copper transporter), member 1
Length=178
Score = 55.5 bits (132), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 34/118 (28%), Positives = 58/118 (49%), Gaps = 6/118 (5%)
Query 41 GLPLPMWFEASSEVLLLFSWWNARTTAQYVVCCLCCIAFGFVSIALKVLRRVSEVRLKMA 100
+ + M F S E ++LF W Y + CL I G ++ + + + +K
Sbjct 42 SIAMQMTFHQSFESVILFESWRTSNRFDYFISCLFIILMGCFTMFISSINKKYIKEIKKN 101
Query 101 EKKHKPTLMLGSLPVFHNALRASVAFLNYSWDYMLMLVAMTFNVGIFLSMLGGMALGF 158
+H+ LG + N L + L Y Y+LML+AMTFN G+F S++ G+++G+
Sbjct 102 RVEHEN---LGIKVICTNVL---LTILYYFMHYLLMLIAMTFNWGLFFSVIIGLSIGY 153
> cel:F58G6.3 hypothetical protein
Length=134
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 34/117 (29%), Positives = 62/117 (52%), Gaps = 4/117 (3%)
Query 46 MWFEASSEVLLLFSWWNARTTAQYVVCCLCCIAFGFVSIALKVLRRVSEVRLKMAEKKHK 105
MWF + +LFS WN + + V C+ G + A+K RR+ + R ++K+
Sbjct 18 MWFHTKPQDTVLFSTWNITSAGKMVWACILVAIAGIILEAIKYNRRLIQKRQSPSKKESY 77
Query 106 PTLMLGSLPVFHNALRASVAFLNYSWDYMLMLVAMTFNVGIFLSMLGGMALGFLTIG 162
+ +L ++ F L F+ + Y LML+ MTF++ + L+++ G+++GFL G
Sbjct 78 ISRLLSTMHFFQTFL----FFVQLGFSYCLMLIFMTFSIWLGLAVVIGLSIGFLIFG 130
> cel:F58G6.7 hypothetical protein; K14686 solute carrier family
31 (copper transporter), member 1
Length=166
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 41/143 (28%), Positives = 59/143 (41%), Gaps = 28/143 (19%)
Query 47 WFEASSEVLLLFSWWNARTTAQYVVCCLCCIAFGFVSIALKVLRRVSEVRLKMAEKK--- 103
+F E +LF W T YV C+ F ALK R ++K+ EKK
Sbjct 9 YFHFRIEEPILFREWKPLNTTAYVFSCIEIFLIAFCLEALKFGRTKLSPKVKIVEKKVDC 68
Query 104 ------------------HKPTLMLGS------LPVFHNALRASVAFLNYSWDYMLMLVA 139
+ T+ L + FH A + + F+ + DY LMLV+
Sbjct 69 CCSTEKDGLWNIPETIPLTQKTVTLAPFTRDSLISKFHMA-SSLLVFVQHFIDYSLMLVS 127
Query 140 MTFNVGIFLSMLGGMALGFLTIG 162
MT+N IFLS+L G G+ +G
Sbjct 128 MTYNWPIFLSLLAGHTTGYFFLG 150
> cel:F58G6.9 hypothetical protein
Length=156
Score = 43.9 bits (102), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 37/134 (27%), Positives = 56/134 (41%), Gaps = 10/134 (7%)
Query 46 MWFEASSEVLLLFSWWNARTTAQYVVCCLCCIAFGFVSIALKVLRRVSEVRLKMAEKKHK 105
MW+ E +LF W V C A G + ALK R +E R+K+ ++
Sbjct 23 MWYHVDVEDTVLFKSWTVFDAGTMVWTCFVVAAAGILLEALKYARWATEERMKIDQENVD 82
Query 106 PTLMLGSLPV------FHNALRASVAFLNYSWD----YMLMLVAMTFNVGIFLSMLGGMA 155
G + + ++ R + L + W Y+LM V M F+V I LS+ G+A
Sbjct 83 SKTKYGGIKIPGKSEKYNFWKRHIIDSLYHFWQLLLAYILMNVYMVFSVYICLSLCFGLA 142
Query 156 LGFLTIGRYLGFSL 169
+G G SL
Sbjct 143 IGHFVFASRTGSSL 156
> ath:AT5G59030 COPT1; COPT1 (copper transporter 1); copper ion
transmembrane transporter; K14686 solute carrier family 31
(copper transporter), member 1
Length=170
Score = 42.4 bits (98), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/107 (27%), Positives = 50/107 (46%), Gaps = 13/107 (12%)
Query 56 LLFSWWNARTTAQYVVCCLCCIAFGFVSIALKVLRRVSEVRLKMAEKKHKPTLMLGSLPV 115
+LFS W ++ Y LC I F+++ + L S +R + ++ ++
Sbjct 54 VLFSGWPGTSSGMY---ALCLIFVFFLAVLTEWLAHSSLLRGSTGDSANRAAGLI----- 105
Query 116 FHNALRASVAFLNYSWDYMLMLVAMTFNVGIFLSMLGGMALGFLTIG 162
+ +V L Y++ML M+FN G+FL L G A+GF+ G
Sbjct 106 -----QTAVYTLRIGLAYLVMLAVMSFNAGVFLVALAGHAVGFMLFG 147
> cel:K12C11.6 hypothetical protein
Length=132
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 37/126 (29%), Positives = 64/126 (50%), Gaps = 15/126 (11%)
Query 44 LPMWFEASSEVLLLFSWWNARTTAQYV-VCCLCCIAFGFVSIALKVLRRVSEVRLKMAEK 102
+ MWF ++ +LF WN T V VCC+ +A I L++++ + R K+ EK
Sbjct 9 MHMWFHTKTQDTVLFKTWNVTDTPTMVWVCCIIVVA----GILLELIKFL---RWKI-EK 60
Query 103 KHKPTLMLGSLPVFHNALR-----ASVAFL-NYSWDYMLMLVAMTFNVGIFLSMLGGMAL 156
HK L S ++ F+ S+ Y+LML+ MTF+V + ++++ G+ +
Sbjct 61 WHKNRDELVSRSYISRLFSPIHIGQTILFMVQLSFSYILMLLFMTFSVWLGIAVVVGLGI 120
Query 157 GFLTIG 162
G+L G
Sbjct 121 GYLAFG 126
> cel:K12C11.7 hypothetical protein
Length=166
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 36/145 (24%), Positives = 59/145 (40%), Gaps = 32/145 (22%)
Query 47 WFEASSEVLLLFSWWNARTTAQYVVCCLCCIAFGFVSIALKVL-----RRVSEVRLKMA- 100
+F + +LF W T YV CI+ F++ L++L R V+ K+A
Sbjct 9 YFHFRIQEPILFRQWKPTDTTGYV---FSCISLFFIAFCLELLKFGRQRMTRTVKEKLAV 65
Query 101 -----------------------EKKHKPTLMLGSLPVFHNALRASVAFLNYSWDYMLML 137
+ + S+ + + + + FL DY LML
Sbjct 66 DCCCSTPEGIWEIPEEPEPSPRGKLASLAPFTMESISSWRHFASSFLFFLQNFVDYSLML 125
Query 138 VAMTFNVGIFLSMLGGMALGFLTIG 162
VAMT+N +F S+L G A+G+ +G
Sbjct 126 VAMTYNYPLFFSLLAGHAIGYFFVG 150
> ath:AT2G26975 copper transporter, putative
Length=145
Score = 40.8 bits (94), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 33/147 (22%), Positives = 70/147 (47%), Gaps = 19/147 (12%)
Query 16 VNHTTVVYSIPTSCVRISSTMLNKCGLPLPMWFEASSEVLLLFSWWNARTTAQYVVCCLC 75
++H + S P+S V +++ N + + ++ ++E+L FS W + YV+C +
Sbjct 1 MDHGNMPPSSPSSMVNHTNS--NMIMMHMTFFWGKNTEIL--FSGWPGTSLGMYVLCLIV 56
Query 76 CIAFGFVSIALKVLRRVSEVRLKMAEKKHKPTLMLGSLPVFHNALRASVAFLNYSWDYML 135
+++ ++ L S +R + + + K ++ +V L Y++
Sbjct 57 VF---LLAVIVEWLAHSSILRGRGSTSRAK------------GLVQTAVYTLKTGLAYLV 101
Query 136 MLVAMTFNVGIFLSMLGGMALGFLTIG 162
ML M+FN G+F+ + G A+GF+ G
Sbjct 102 MLAVMSFNGGVFIVAIAGFAVGFMLFG 128
> cel:Y58A7A.1 hypothetical protein
Length=130
Score = 39.7 bits (91), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 25/108 (23%), Positives = 45/108 (41%), Gaps = 2/108 (1%)
Query 46 MWFEASSEVLLLFSWWNARTTAQYVVCCLCCIAFGFVSIALKVLRRVSEVRLKMAEKKHK 105
M F +E +LF +W T V C + F+ L+ R + + ++ +
Sbjct 1 MSFHFGTEETILFDFWKTETAVGIAVACFITVLLAFLMETLRFFRDYRKAQTQLHQPPIS 60
Query 106 PTLMLGSLPVFHNALRASVAFLNYSWDYMLMLVAMTFNVGI-FLSMLG 152
P L P + + + + Y LML+ MTFN + F +++G
Sbjct 61 PEDRLKRSPQL-DLIDPLLQLFQLTIAYFLMLIFMTFNAYLCFFTVVG 107
> ath:AT5G20650 COPT5; COPT5; copper ion transmembrane transporter/
high affinity copper ion transmembrane transporter; K14686
solute carrier family 31 (copper transporter), member 1
Length=146
Score = 38.5 bits (88), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 29/116 (25%), Positives = 54/116 (46%), Gaps = 11/116 (9%)
Query 53 EVLLLFSWWNARTTAQYVVCCLCCIAFGFVSIALKVLRRVSEVRLKMAEKKHKPTLMLG- 111
+ +LF +W + Y++ + C F L+ RR+ L + + P
Sbjct 11 KATILFDFWKTDSWLSYILTLIACFVFSAFYQYLEN-RRIQFKSLSSSRRAPPPPRSSSG 69
Query 112 -SLPVF-----HNALRASVAFL---NYSWDYMLMLVAMTFNVGIFLSMLGGMALGF 158
S P+ +A +A+ L N + Y+LML AM+FN G+F++++ G+ G+
Sbjct 70 VSAPLIPKSGTRSAAKAASVLLFGVNAAIGYLLMLAAMSFNGGVFIAIVVGLTAGY 125
> ath:AT5G59040 COPT3; COPT3; copper ion transmembrane transporter/
high affinity copper ion transmembrane transporter
Length=151
Score = 37.4 bits (85), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 31/125 (24%), Positives = 51/125 (40%), Gaps = 16/125 (12%)
Query 38 NKCGLPLPMWFEASSEVLLLFSWWNARTTAQYVVCCLCCIAFGFVSIALKVLRRVSEVRL 97
++ G + M F +LF W + Y VC V+ SE
Sbjct 24 HRHGGMMHMTFFWGKTTEVLFDGWPGTSLKMYWVCLAVIF----------VISAFSECLS 73
Query 98 KMAEKKHKPTLMLGSLPVFHNALRASVAFLNYSWDYMLMLVAMTFNVGIFLSMLGGMALG 157
+ K P + G L L+ +V + + Y++ML M+FN G+F++ + G LG
Sbjct 74 RCGFMKSGPASLGGGL------LQTAVYTVRAALSYLVMLAVMSFNGGVFVAAMAGFGLG 127
Query 158 FLTIG 162
F+ G
Sbjct 128 FMIFG 132
> tgo:TGME49_021350 hypothetical protein
Length=300
Score = 36.6 bits (83), Expect = 0.091, Method: Compositional matrix adjust.
Identities = 15/26 (57%), Positives = 21/26 (80%), Gaps = 0/26 (0%)
Query 132 DYMLMLVAMTFNVGIFLSMLGGMALG 157
D+ LMLV MTFN G+FL+++ G+A G
Sbjct 249 DWSLMLVCMTFNAGLFLTVVAGVAAG 274
> ath:AT3G46900 COPT2; COPT2; copper ion transmembrane transporter/
high affinity copper ion transmembrane transporter
Length=158
Score = 36.6 bits (83), Expect = 0.092, Method: Compositional matrix adjust.
Identities = 28/108 (25%), Positives = 46/108 (42%), Gaps = 17/108 (15%)
Query 56 LLFSWWNARTTAQYVVCCLCCIAFGFVSIALKVLRRVSEVRLKMAEKKHKPTLML-GSLP 114
+LFS W ++ Y +C I + +L ++E H P L + GS
Sbjct 42 VLFSGWPGTSSGMYALCL----------IVIFLLAVIAE------WLAHSPILRVSGSTN 85
Query 115 VFHNALRASVAFLNYSWDYMLMLVAMTFNVGIFLSMLGGMALGFLTIG 162
+ +V L Y++ML M+FN G+F+ + G +GF G
Sbjct 86 RAAGLAQTAVYTLKTGLSYLVMLAVMSFNAGVFIVAIAGYGVGFFLFG 133
> tgo:TGME49_025260 DNA-directed RNA polymerase II largest subunit,
putative (EC:2.7.7.6 3.5.1.16); K03006 DNA-directed RNA
polymerase II subunit RPB1 [EC:2.7.7.6]
Length=1892
Score = 34.7 bits (78), Expect = 0.35, Method: Composition-based stats.
Identities = 28/91 (30%), Positives = 42/91 (46%), Gaps = 7/91 (7%)
Query 40 CGLPLPMWFEASSEVLLLFSWWNARTTAQYVVCCLCCIAFGFVSIALKVLRRVSEVRLKM 99
CG P +F+ +++LF + R V F A VLRR+SE LKM
Sbjct 174 CGCVQPRYFKEGPNIMVLFP--DNREEGDEDVTEDIRRIFA-AEEAYAVLRRISEEDLKM 230
Query 100 ----AEKKHKPTLMLGSLPVFHNALRASVAF 126
E+ H + +L +LP+ A+R SV +
Sbjct 231 MGFDPERAHPASFILSTLPIPPLAVRPSVQY 261
> sce:YPR124W CTR1; Ctr1p
Length=406
Score = 34.3 bits (77), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 32/58 (55%), Gaps = 4/58 (6%)
Query 106 PTLMLGSLPVFHNALRASVAFLNYSWDYMLMLVAMTFNVGIFLSMLGGMALGFLTIGR 163
P+LM +FH+ +RA + F + YMLML M+F + +++ G+AL + R
Sbjct 230 PSLM----DLFHDIIRAFLVFTSTMIIYMLMLATMSFVLTYVFAVITGLALSEVFFNR 283
> dre:563012 sb:cb797; si:ch211-282j22.4; K14687 solute carrier
family 31 (copper transporter), member 2
Length=171
Score = 33.5 bits (75), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 25/40 (62%), Gaps = 0/40 (0%)
Query 120 LRASVAFLNYSWDYMLMLVAMTFNVGIFLSMLGGMALGFL 159
L+ ++ + + YMLML M++N+ IFL ++ G LG+
Sbjct 123 LQTAIHIVQVTLGYMLMLCVMSYNIWIFLGVITGSVLGYF 162
> sce:YHR175W CTR2; Ctr2p; K14686 solute carrier family 31 (copper
transporter), member 1
Length=189
Score = 33.1 bits (74), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 27/107 (25%), Positives = 49/107 (45%), Gaps = 14/107 (13%)
Query 56 LLFSWWNARTTAQYVVCCLCCIAFGFVSIALK--VLRRVSEVRLKMAEKKHKPTLMLGSL 113
++F WW+ +T ++ CL ++ LK V +R R+ + + SL
Sbjct 72 VVFEWWHIKTLPGLILSCLAIFGLAYLYEYLKYCVHKRQLSQRVLLPNR---------SL 122
Query 114 PVFHNALRASVAFL---NYSWDYMLMLVAMTFNVGIFLSMLGGMALG 157
+ A + S + L + +MLMLV MT+N + L+++ G G
Sbjct 123 TKINQADKVSNSILYGLQVGFSFMLMLVFMTYNGWLMLAVVCGAIWG 169
> mmu:13733 Emr1, DD7A5-7, EGF-TM7, F4/80, Gpf480, Ly71, TM7LN3;
EGF-like module containing, mucin-like, hormone receptor-like
sequence 1; K04591 egf-like module containing, mucin-like,
hormone receptor-like 1
Length=931
Score = 32.3 bits (72), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 22/76 (28%), Positives = 33/76 (43%), Gaps = 15/76 (19%)
Query 170 QPKNIVSGCECDEDFSC----------GCHK---GQSCTCCVSSQLDPANGGPSISGKDP 216
QP +++G C+++ C CH CTC +S L+ + GGP G D
Sbjct 161 QPGFVLNGSICEDEDECVTRDVCPEHATCHNTLGSYYCTC--NSGLESSGGGPMFQGLDE 218
Query 217 VCGHSGQCKTYRTVCG 232
C +C T+CG
Sbjct 219 SCEDVDECSRNSTLCG 234
> dre:100317326 si:ch1073-55a19.2; K12172 E3 SUMO-protein ligase
RanBP2
Length=2950
Score = 32.3 bits (72), Expect = 1.7, Method: Composition-based stats.
Identities = 15/31 (48%), Positives = 22/31 (70%), Gaps = 4/31 (12%)
Query 77 IAFGFVSIALKVLRRVSEVRLKMAEKKHKPT 107
+AFGFV+ ++VLRR++E M K+ KPT
Sbjct 2913 VAFGFVTDGMQVLRRLAE----MGTKEGKPT 2939
> mmu:16773 Lama2, 5830440B04, KIAA4087, dy, mKIAA4087, mer, merosin;
laminin, alpha 2; K05637 laminin, alpha 1/2
Length=3118
Score = 32.0 bits (71), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 35/156 (22%), Positives = 65/156 (41%), Gaps = 18/156 (11%)
Query 84 IALKVLRRVSEVRLKMAEKKHKPTLMLGSLPVFHNALRASVAFLNYSWDYMLMLVAMTFN 143
+A ++ +++ ++M EK+ K G P + FL+ +D +L+ T+
Sbjct 1287 MAAPLIGQLTRHEIEMTEKEWK---YYGDDPRISRTV-TREDFLDILYDIHYILIKATYG 1342
Query 144 VGIFLSMLGGMALGFLTIGRYLGFSLQPKNIVSGCECDEDFSCGCHKGQSCTCC------ 197
+ S + +++ G L S P +++ C+C +S G SC C
Sbjct 1343 NVVRQSRISEISMEVAEPGHVLAGS-PPAHLIERCDCPPGYS-----GLSCETCAPGFYR 1396
Query 198 VSSQLDPANGGPSISGKDPV--CGHSGQCKTYRTVC 231
+ S+ GP++ P GHS QC +VC
Sbjct 1397 LRSEPGGRTPGPTLGTCVPCQCNGHSSQCDPETSVC 1432
> cel:R08F11.1 hypothetical protein
Length=884
Score = 31.6 bits (70), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 29/61 (47%), Gaps = 3/61 (4%)
Query 71 VCCLCCIAFGFVSIALKVLRRVSEVRLKMAEKKHKPTLMLGSLPV---FHNALRASVAFL 127
+ CL C FG +S+ LK+L + +K +E HK + G V N L S+A
Sbjct 808 IFCLLCNNFGKISMKLKLLYLFVAITIKFSEFSHKIRVFSGKKLVSSKSENVLNISIALP 867
Query 128 N 128
N
Sbjct 868 N 868
> cel:F31E8.4 hypothetical protein; K14686 solute carrier family
31 (copper transporter), member 1
Length=162
Score = 31.6 bits (70), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 21/27 (77%), Gaps = 0/27 (0%)
Query 133 YMLMLVAMTFNVGIFLSMLGGMALGFL 159
Y LML+AMT+N+ + LS++ G A+G+
Sbjct 120 YTLMLIAMTYNMNLILSIVVGEAVGYF 146
> xla:397694 slc31a2, copt2, ctr2, xem1; solute carrier family
31 (copper transporters), member 2; K14687 solute carrier family
31 (copper transporter), member 2
Length=172
Score = 31.2 bits (69), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 38/77 (49%), Gaps = 9/77 (11%)
Query 96 RLKMAEKKHKPTLMLGSLPVFHNALRASVAFLNYSWDYMLMLVAMTFNVGIFLSMLGGMA 155
RL + E+ +P+ L F + LR L Y +LML M++N IF++++ G
Sbjct 98 RLSVTEEHIQPSSRWWFLHSFLSLLRMVQVVLGY----LLMLCVMSYNAAIFIAVILGSG 153
Query 156 LGFLTIGRYLGFSLQPK 172
LG+ +L F L K
Sbjct 154 LGY-----FLAFPLLSK 165
> ath:AT2G37925 COPT4; COPT4; copper ion transmembrane transporter/
high affinity copper ion transmembrane transporter
Length=145
Score = 30.8 bits (68), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 25/103 (24%), Positives = 46/103 (44%), Gaps = 13/103 (12%)
Query 56 LLFSWWNARTTAQYVVCCLCCIAFGFVSIALKVLRRVSEVRLKMAEKKHKPTLMLGSLPV 115
+LFS W Y + + F++ L S ++ + A+K K
Sbjct 42 VLFSGWPGSDRGMYALALIFVFFLAFLAEWLARCSDASSIK-QGADKLAKV--------- 91
Query 116 FHNALRASVAFLNYSWDYMLMLVAMTFNVGIFLSMLGGMALGF 158
A R ++ + + Y+++L ++FN G+FL+ + G ALGF
Sbjct 92 ---AFRTAMYTVKSGFSYLVILAVVSFNGGVFLAAIFGHALGF 131
> cel:K12C11.3 hypothetical protein
Length=147
Score = 30.4 bits (67), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 29/128 (22%), Positives = 55/128 (42%), Gaps = 9/128 (7%)
Query 47 WFEASSEVLLLFSWWNARTTAQYVVCCLCCIAFGFVSIALKVLRRVSEVRLKMA---EKK 103
W+ ++LF W + + C GF+ LK + + ++++ A +++
Sbjct 20 WYHVELNDVILFENWKVQDMTTMIWSCFVVGFAGFLLEFLKYSKWAASMQMRPAGDVDRR 79
Query 104 HK------PTLMLGSLPVFHNALRASVAFLNYSWDYMLMLVAMTFNVGIFLSMLGGMALG 157
K P+ L + ++A F ++LM + MTFNV I LS+ G+ +G
Sbjct 80 TKYGGCVVPSENRKKLFWARHVVQAMYHFWQTLLAFILMNIYMTFNVYICLSLCLGLTIG 139
Query 158 FLTIGRYL 165
+ G L
Sbjct 140 YFFFGSRL 147
Lambda K H
0.325 0.137 0.453
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8338372120
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40