bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0993_orf2
Length=102
Score E
Sequences producing significant alignments: (Bits) Value
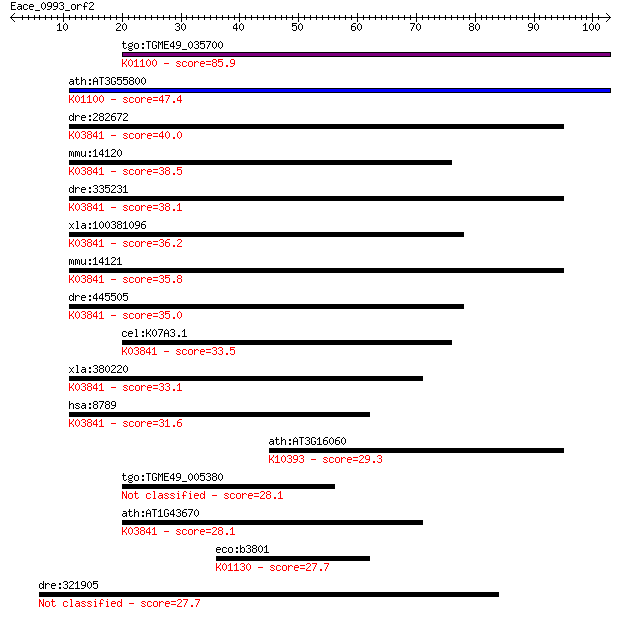
tgo:TGME49_035700 sedoheptulose-1,7 bisphosphatase, putative (... 85.9 3e-17
ath:AT3G55800 SBPASE; SBPASE (sedoheptulose-bisphosphatase); p... 47.4 1e-05
dre:282672 fbp1b, cb598, fb57b01, fbp1, id:ibd1091, wu:fb17g10... 40.0 0.002
mmu:14120 Fbp2, Fbp-1, Fbp1, Rae-30; fructose bisphosphatase 2... 38.5 0.006
dre:335231 fbp1a, fbp1, fbp1l, fk92h02, wu:fk92h02, zgc:64096;... 38.1 0.006
xla:100381096 fbp2; fructose-1,6-bisphosphatase 2 (EC:3.1.3.11... 36.2 0.026
mmu:14121 Fbp1, Fbp-2, Fbp2, Fbp3; fructose bisphosphatase 1 (... 35.8 0.037
dre:445505 fbp2, zgc:101083; fructose-1,6-bisphosphatase 2 (EC... 35.0 0.051
cel:K07A3.1 fbp-1; Fructose-1,6-BiPhosphatase family member (f... 33.5 0.18
xla:380220 fbp1, MGC64343, fbp; fructose-1,6-bisphosphatase 1 ... 33.1 0.22
hsa:8789 FBP2, MGC142192; fructose-1,6-bisphosphatase 2 (EC:3.... 31.6 0.71
ath:AT3G16060 kinesin motor family protein; K10393 kinesin fam... 29.3 3.2
tgo:TGME49_005380 fructose-1,6-bisphosphatase, putative (EC:3.... 28.1 6.3
ath:AT1G43670 fructose-1,6-bisphosphatase, putative / D-fructo... 28.1 6.4
eco:b3801 aslA, atsA, ECK3794, gppB, JW3773; acrylsulfatase-li... 27.7 8.1
dre:321905 phrf1, fb38b02, fc78a10, wu:fb38b02, wu:fc78a10, zg... 27.7 9.0
> tgo:TGME49_035700 sedoheptulose-1,7 bisphosphatase, putative
(EC:3.1.3.37); K01100 sedoheptulose-bisphosphatase [EC:3.1.3.37]
Length=330
Score = 85.9 bits (211), Expect = 3e-17, Method: Composition-based stats.
Identities = 46/90 (51%), Positives = 63/90 (70%), Gaps = 8/90 (8%)
Query 20 GRQLVAAVLVIYGPRTTALLS----AGGSTFD---LQLDPEEGDPVVLRGPCSIQKEGAK 72
GRQ VA+++V+YGPRTT +++ AGG + L L+ ++ + RG I K AK
Sbjct 148 GRQQVASLIVVYGPRTTGVVAVNVDAGGIVKEGTALDLEMKDNGKFICRGK-PIIKPQAK 206
Query 73 IFSPANLRAAQDLPAYNRMVQRWMEKRFTL 102
IFSPANLRAAQDLPAY ++++ WMEKR+TL
Sbjct 207 IFSPANLRAAQDLPAYKQLIEFWMEKRYTL 236
> ath:AT3G55800 SBPASE; SBPASE (sedoheptulose-bisphosphatase);
phosphoric ester hydrolase/ sedoheptulose-bisphosphatase (EC:3.1.3.37);
K01100 sedoheptulose-bisphosphatase [EC:3.1.3.37]
Length=393
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 32/94 (34%), Positives = 51/94 (54%), Gaps = 5/94 (5%)
Query 11 PNDYNASPMGRQLVAAVLVIYGPRTTALLSAGG--STFDLQLDPEEGDPVVLRGPCSIQK 68
P D G VAA + IYGPRTT +L+ G T + L +EG ++ I +
Sbjct 199 PGDKLTGITGGDQVAAAMGIYGPRTTYVLAVKGFPGTHEFLL-LDEGKWQHVKETTEIAE 257
Query 69 EGAKIFSPANLRAAQDLPAYNRMVQRWMEKRFTL 102
K+FSP NLRA D Y++++ ++++++TL
Sbjct 258 --GKMFSPGNLRATFDNSEYSKLIDYYVKEKYTL 289
> dre:282672 fbp1b, cb598, fb57b01, fbp1, id:ibd1091, wu:fb17g10,
wu:fb57b01, zgc:64127; fructose-1,6-bisphosphatase 1b (EC:3.1.3.11);
K03841 fructose-1,6-bisphosphatase I [EC:3.1.3.11]
Length=337
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 27/84 (32%), Positives = 42/84 (50%), Gaps = 1/84 (1%)
Query 11 PNDYNASPMGRQLVAAVLVIYGPRTTALLSAGGSTFDLQLDPEEGDPVVLRGPCSIQKEG 70
P++ +A GR +VAA +YG T +LS G LDP G+ +++ I+K+G
Sbjct 148 PSEKDALRSGRHIVAAGYALYGSATMLVLSTGQGVNCFMLDPAIGEFILVDRDVRIKKKG 207
Query 71 AKIFSPANLRAAQDLPAYNRMVQR 94
KI+S A Q P +Q+
Sbjct 208 -KIYSLNEGYAQQFYPDVTEYLQK 230
> mmu:14120 Fbp2, Fbp-1, Fbp1, Rae-30; fructose bisphosphatase
2 (EC:3.1.3.11); K03841 fructose-1,6-bisphosphatase I [EC:3.1.3.11]
Length=339
Score = 38.5 bits (88), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 35/65 (53%), Gaps = 1/65 (1%)
Query 11 PNDYNASPMGRQLVAAVLVIYGPRTTALLSAGGSTFDLQLDPEEGDPVVLRGPCSIQKEG 70
P++ +A GR +VAA +YG T LS G LDP G+ V++ I+K+G
Sbjct 148 PSEKDALQPGRNIVAAGYALYGSATLVALSTGQGVDLFMLDPALGEFVLVEKDVRIKKKG 207
Query 71 AKIFS 75
KIFS
Sbjct 208 -KIFS 211
> dre:335231 fbp1a, fbp1, fbp1l, fk92h02, wu:fk92h02, zgc:64096;
fructose-1,6-bisphosphatase 1a; K03841 fructose-1,6-bisphosphatase
I [EC:3.1.3.11]
Length=337
Score = 38.1 bits (87), Expect = 0.006, Method: Composition-based stats.
Identities = 27/84 (32%), Positives = 42/84 (50%), Gaps = 1/84 (1%)
Query 11 PNDYNASPMGRQLVAAVLVIYGPRTTALLSAGGSTFDLQLDPEEGDPVVLRGPCSIQKEG 70
P++ +A GR +VAA +YG T +LS G LDP G+ +++ I+K+G
Sbjct 148 PSEKDALQPGRNIVAAGYALYGSATMIVLSTGQGVNCFMLDPAIGEFILVDRDVKIKKKG 207
Query 71 AKIFSPANLRAAQDLPAYNRMVQR 94
KI+S A PA +Q+
Sbjct 208 -KIYSLNEGYALYFDPAVTEYLQK 230
> xla:100381096 fbp2; fructose-1,6-bisphosphatase 2 (EC:3.1.3.11);
K03841 fructose-1,6-bisphosphatase I [EC:3.1.3.11]
Length=339
Score = 36.2 bits (82), Expect = 0.026, Method: Composition-based stats.
Identities = 26/76 (34%), Positives = 35/76 (46%), Gaps = 9/76 (11%)
Query 11 PNDYNASPMGRQLVAAVLVIYGPRTTALLSAGGSTFDLQLDPEEGDPVVLRGPCSIQKEG 70
P + +A GR +VAA +YG T LS G LDP G+ V++ I+K+G
Sbjct 148 PCEKDALQPGRNIVAAGYALYGSATLVALSTGQGVDCFMLDPALGEFVLVDKNVRIKKKG 207
Query 71 ---------AKIFSPA 77
AK F PA
Sbjct 208 KIYSLNEGYAKYFDPA 223
> mmu:14121 Fbp1, Fbp-2, Fbp2, Fbp3; fructose bisphosphatase 1
(EC:3.1.3.11); K03841 fructose-1,6-bisphosphatase I [EC:3.1.3.11]
Length=338
Score = 35.8 bits (81), Expect = 0.037, Method: Composition-based stats.
Identities = 28/85 (32%), Positives = 44/85 (51%), Gaps = 3/85 (3%)
Query 11 PNDYNASPMGRQLVAAVLVIYGPRTTALLSAGGSTFDLQLDPEEGDPVVLRGPCSIQKEG 70
P++ +A GR LVAA +YG T +L+ LDP G+ +++ ++K+G
Sbjct 148 PSEKDALQPGRDLVAAGYALYGSATMLVLAMDCGVNCFMLDPSIGEFIMVDRDVKMKKKG 207
Query 71 AKIFSPANLRAAQDL-PAYNRMVQR 94
I+S N A+D PA N +QR
Sbjct 208 -NIYS-LNEGYAKDFDPAINEYLQR 230
> dre:445505 fbp2, zgc:101083; fructose-1,6-bisphosphatase 2 (EC:3.1.3.11);
K03841 fructose-1,6-bisphosphatase I [EC:3.1.3.11]
Length=338
Score = 35.0 bits (79), Expect = 0.051, Method: Composition-based stats.
Identities = 24/76 (31%), Positives = 34/76 (44%), Gaps = 9/76 (11%)
Query 11 PNDYNASPMGRQLVAAVLVIYGPRTTALLSAGGSTFDLQLDPEEGDPVVLRGPCSIQKEG 70
P++ +A G Q+V A +YG T LS G LDP G+ ++ I+K+G
Sbjct 149 PSEKDALQPGNQIVCAGYALYGSATLVALSTGAGVNFFMLDPAIGEFILTDRNVKIKKKG 208
Query 71 ---------AKIFSPA 77
AK F PA
Sbjct 209 KTYSINEGYAKYFEPA 224
> cel:K07A3.1 fbp-1; Fructose-1,6-BiPhosphatase family member
(fbp-1); K03841 fructose-1,6-bisphosphatase I [EC:3.1.3.11]
Length=341
Score = 33.5 bits (75), Expect = 0.18, Method: Composition-based stats.
Identities = 20/56 (35%), Positives = 28/56 (50%), Gaps = 1/56 (1%)
Query 20 GRQLVAAVLVIYGPRTTALLSAGGSTFDLQLDPEEGDPVVLRGPCSIQKEGAKIFS 75
G+++VAA +YG T +LS G LDP G+ +L P KE I+S
Sbjct 161 GKEMVAAGYALYGSATMVVLSTGDGVNGFTLDPSIGE-FILTHPNMKCKEKGSIYS 215
> xla:380220 fbp1, MGC64343, fbp; fructose-1,6-bisphosphatase
1 (EC:3.1.3.11); K03841 fructose-1,6-bisphosphatase I [EC:3.1.3.11]
Length=338
Score = 33.1 bits (74), Expect = 0.22, Method: Composition-based stats.
Identities = 19/60 (31%), Positives = 31/60 (51%), Gaps = 0/60 (0%)
Query 11 PNDYNASPMGRQLVAAVLVIYGPRTTALLSAGGSTFDLQLDPEEGDPVVLRGPCSIQKEG 70
P + +A GR +VAA +YG T +L+A LDP G+ +++ I+K+G
Sbjct 148 PTEKDALQPGRNIVAAGYALYGSATMLVLAAECGVNCFMLDPAIGEFILVNQDVKIKKKG 207
> hsa:8789 FBP2, MGC142192; fructose-1,6-bisphosphatase 2 (EC:3.1.3.11);
K03841 fructose-1,6-bisphosphatase I [EC:3.1.3.11]
Length=339
Score = 31.6 bits (70), Expect = 0.71, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 11 PNDYNASPMGRQLVAAVLVIYGPRTTALLSAGGSTFDLQLDPEEGDPVVLR 61
P++ +A GR +VAA +YG T LS G LDP G+ V++
Sbjct 148 PSEKDALQCGRNIVAAGYALYGSATLVALSTGQGVDLFMLDPALGEFVLVE 198
> ath:AT3G16060 kinesin motor family protein; K10393 kinesin family
member 2/24
Length=684
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 25/50 (50%), Gaps = 8/50 (16%)
Query 45 TFDLQLDPEEGDPVVLRGPCSIQKEGAKIFSPANLRAAQDLPAYNRMVQR 94
+FD +L PE P S Q +G +F+P+ ++ D AYN+ R
Sbjct 108 SFDTELLPEI--------PVSNQLDGPSLFNPSQGQSFDDFEAYNKQPNR 149
> tgo:TGME49_005380 fructose-1,6-bisphosphatase, putative (EC:3.1.3.11)
Length=388
Score = 28.1 bits (61), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 12/36 (33%), Positives = 18/36 (50%), Gaps = 0/36 (0%)
Query 20 GRQLVAAVLVIYGPRTTALLSAGGSTFDLQLDPEEG 55
G++ V A V+YG T +++ G LDP G
Sbjct 187 GKEQVCAGYVLYGSSTVMVITVGSGVHGFTLDPSLG 222
> ath:AT1G43670 fructose-1,6-bisphosphatase, putative / D-fructose-1,6-bisphosphate
1-phosphohydrolase, putative / FBPase,
putative (EC:3.1.3.11); K03841 fructose-1,6-bisphosphatase
I [EC:3.1.3.11]
Length=341
Score = 28.1 bits (61), Expect = 6.4, Method: Composition-based stats.
Identities = 15/51 (29%), Positives = 23/51 (45%), Gaps = 0/51 (0%)
Query 20 GRQLVAAVLVIYGPRTTALLSAGGSTFDLQLDPEEGDPVVLRGPCSIQKEG 70
G ++VAA +YG +LS G LDP G+ ++ I +G
Sbjct 159 GNEMVAAGYCMYGSSCMLVLSTGTGVHGFTLDPSLGEFILTHPDIKIPNKG 209
> eco:b3801 aslA, atsA, ECK3794, gppB, JW3773; acrylsulfatase-like
enzyme (EC:3.1.6.1); K01130 arylsulfatase [EC:3.1.6.1]
Length=551
Score = 27.7 bits (60), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 11/26 (42%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 36 TALLSAGGSTFDLQLDPEEGDPVVLR 61
T + +AG S F+L DP+E D + +R
Sbjct 495 TVMQTAGSSVFNLYTDPQESDSIGVR 520
> dre:321905 phrf1, fb38b02, fc78a10, wu:fb38b02, wu:fc78a10,
zgc:172184; PHD and ring finger domains 1
Length=944
Score = 27.7 bits (60), Expect = 9.0, Method: Composition-based stats.
Identities = 20/78 (25%), Positives = 34/78 (43%), Gaps = 11/78 (14%)
Query 6 PAAYSPNDYNASPMGRQLVAAVLVIYGPRTTALLSAGGSTFDLQLDPEEGDPVVLRGPCS 65
P+ Y P++Y+ M ++ AA L +Y G FDL + D + + P S
Sbjct 401 PSVYRPSEYSLGSMRAEIGAASLSVY-----------GDPFDLDPFNDREDEIQVLTPSS 449
Query 66 IQKEGAKIFSPANLRAAQ 83
+ + S + LR+ Q
Sbjct 450 VLDAKRRGLSRSALRSHQ 467
Lambda K H
0.319 0.135 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2031832220
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40