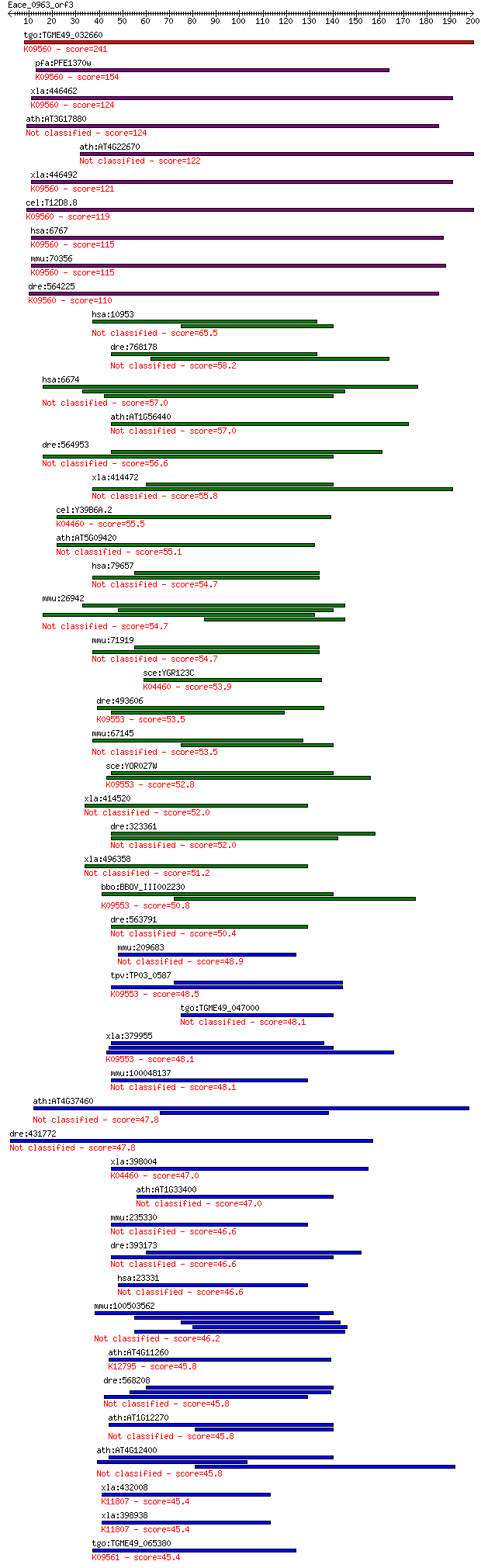
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0963_orf3
Length=199
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_032660 58 kDa phosphoprotein, putative ; K09560 sup... 241 2e-63
pfa:PFE1370w hsp70 interacting protein, putative; K09560 suppr... 154 3e-37
xla:446462 st13, MGC78939; suppression of tumorigenicity 13 (c... 124 1e-28
ath:AT3G17880 ATTDX; ATTDX (TETRATICOPEPTIDE DOMAIN-CONTAINING... 124 2e-28
ath:AT4G22670 AtHip1; AtHip1 (Arabidopsis thaliana Hsp70-inter... 122 1e-27
xla:446492 MGC79131 protein; K09560 suppressor of tumorigenici... 121 1e-27
cel:T12D8.8 hypothetical protein; K09560 suppressor of tumorig... 119 5e-27
hsa:6767 ST13, AAG2, FAM10A1, FAM10A4, FLJ27260, HIP, HOP, HSP... 115 7e-26
mmu:70356 St13, 1110007I03Rik, 3110002K08Rik, AW555194, HIP, H... 115 1e-25
dre:564225 st13, MGC73267, MGC77089, wu:fd15g02, zgc:73267; su... 110 3e-24
hsa:10953 TOMM34, HTOM34P, TOM34, URCC3; translocase of outer ... 65.5 1e-10
dre:768178 zgc:153288 58.2 2e-08
hsa:6674 SPAG1, FLJ32920, HSD-3.8, SP75, TPIS; sperm associate... 57.0 4e-08
ath:AT1G56440 serine/threonine protein phosphatase-related 57.0 5e-08
dre:564953 spag1, MGC162178, cb1089, wu:fj78g10; sperm associa... 56.6 6e-08
xla:414472 rpap3, MGC81126; RNA polymerase II associated prote... 55.8 9e-08
cel:Y39B6A.2 pph-5; Protein PHosphatase family member (pph-5);... 55.5 1e-07
ath:AT5G09420 ATTOC64-V (ARABIDOPSIS THALIANA TRANSLOCON AT TH... 55.1 1e-07
hsa:79657 RPAP3, FLJ21908; RNA polymerase II associated protein 3 54.7
mmu:26942 Spag1, tpis; sperm associated antigen 1 54.7
mmu:71919 Rpap3, 2310042P20Rik, D15Ertd682e; RNA polymerase II... 54.7 2e-07
sce:YGR123C PPT1; Ppt1p (EC:3.1.3.16); K04460 protein phosphat... 53.9 3e-07
dre:493606 stip1, zgc:92133; stress-induced-phosphoprotein 1 (... 53.5 5e-07
mmu:67145 Tomm34, 2610100K07Rik, TOM34, Tomm34a, Tomm34b; tran... 53.5 5e-07
sce:YOR027W STI1; Hsp90 cochaperone, interacts with the Ssa gr... 52.8 9e-07
xla:414520 hypothetical protein MGC81394 52.0 1e-06
dre:323361 tomm34, wu:fb96b08, zgc:56645; translocase of outer... 52.0 1e-06
xla:496358 sgta; small glutamine-rich tetratricopeptide repeat... 51.2 2e-06
bbo:BBOV_III002230 17.m07215; tetratricopeptide repeat domain ... 50.8 3e-06
dre:563791 ttc12; tetratricopeptide repeat domain 12 50.4
mmu:209683 Ttc28, 2310015L07Rik, 6030435N04, AI428795, AI85176... 48.9 1e-05
tpv:TP03_0587 hypothetical protein; K09553 stress-induced-phos... 48.5 1e-05
tgo:TGME49_047000 TPR domain-containing protein (EC:3.4.21.72) 48.1 2e-05
xla:379955 stip1, MGC53256; stress-induced-phosphoprotein 1; K... 48.1 2e-05
mmu:100048137 tetratricopeptide repeat protein 12-like 48.1 2e-05
ath:AT4G37460 SRFR1; SRFR1 (SUPPRESSOR OF RPS4-RLD 1); protein... 47.8 2e-05
dre:431772 sgta, zgc:92462; small glutamine-rich tetratricopep... 47.8 3e-05
xla:398004 ppp5c, pp5; protein phosphatase 5, catalytic subuni... 47.0 4e-05
ath:AT1G33400 tetratricopeptide repeat (TPR)-containing protein 47.0 4e-05
mmu:235330 Ttc12, E330017O07Rik; tetratricopeptide repeat doma... 46.6 5e-05
dre:393173 MGC56178; zgc:56178 46.6 5e-05
hsa:23331 TTC28, KIAA1043; tetratricopeptide repeat domain 28 46.6
mmu:100503562 hypothetical protein LOC100503562 46.2 7e-05
ath:AT4G11260 SGT1B; SGT1B; protein binding; K12795 suppressor... 45.8 9e-05
dre:568208 ttc6; tetratricopeptide repeat domain 6 45.8
ath:AT1G12270 stress-inducible protein, putative 45.8 1e-04
ath:AT4G12400 stress-inducible protein, putative 45.8 1e-04
xla:432008 wdtc1, MGC78868; WD and tetratricopeptide repeats 1... 45.4 1e-04
xla:398938 hypothetical protein MGC68614; K11807 WD and tetrat... 45.4 1e-04
tgo:TGME49_065380 TPR domain-containing protein ; K09561 STIP1... 45.4 1e-04
> tgo:TGME49_032660 58 kDa phosphoprotein, putative ; K09560 suppressor
of tumorigenicity protein 13
Length=425
Score = 241 bits (614), Expect = 2e-63, Method: Compositional matrix adjust.
Identities = 131/192 (68%), Positives = 160/192 (83%), Gaps = 0/192 (0%)
Query 8 SLNDEEVVVPESTPAPPLAPAEERELTDEDYEKLAKAKEAATEAAEAGDLNRAVESYTEA 67
SL D EV+ PE++P PPLAP E+ELTD++ +KL K KE A+ A EAG+ RA+E +TEA
Sbjct 85 SLKDSEVIPPETSPLPPLAPEGEKELTDDELDKLGKLKEEASAACEAGNSERALEKFTEA 144
Query 68 LLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQ 127
LL+GNPTALLYTRRADVLLK KR A IRDCDEALKLNPD+ARAY+IRG ANR LG W++
Sbjct 145 LLIGNPTALLYTRRADVLLKLKRPVACIRDCDEALKLNPDSARAYKIRGKANRLLGKWRE 204
Query 128 AHSDIEMGQKIDYDENIWDIQKLVEQKYKIIEEHERAAQRKKEEKERKERERRAREQRAN 187
AHSD++MGQKIDYDE +WD+QKLV++K+K IEEHER RK EE E+K RE+ AR++RA
Sbjct 205 AHSDLDMGQKIDYDEGLWDMQKLVDEKFKKIEEHERKIVRKCEEAEKKRREKEARKRRAA 264
Query 188 AQRAYEEQKKRE 199
AQRAYEEQK+R+
Sbjct 265 AQRAYEEQKQRD 276
> pfa:PFE1370w hsp70 interacting protein, putative; K09560 suppressor
of tumorigenicity protein 13
Length=458
Score = 154 bits (388), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 73/151 (48%), Positives = 102/151 (67%), Gaps = 0/151 (0%)
Query 13 EVVVPESTPAPPLAPAEERELTDEDYEKLAKAKEAATEAAEAGDLNRAVESYTEALLVGN 72
+ +V E+ PPLAP + +L+DE E+++ K A E + A+E Y + + G
Sbjct 102 DFMVEETIECPPLAPIVDEDLSDEVLEEISNLKIEAAELVQDNKFEEALEKYNKIIAFGK 161
Query 73 PTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSDI 132
P+A++YT+RA VLL KR A IRDC EAL LN D+A AY++R A R+LG W+ AH+DI
Sbjct 162 PSAMIYTKRASVLLSLKRPKACIRDCTEALNLNIDSANAYKVRAKAYRHLGKWECAHADI 221
Query 133 EMGQKIDYDENIWDIQKLVEQKYKIIEEHER 163
E GQKIDYDE++W++QKL+E+KYK I E R
Sbjct 222 EQGQKIDYDEDLWEMQKLIEEKYKKIYEKRR 252
> xla:446462 st13, MGC78939; suppression of tumorigenicity 13
(colon carcinoma) (Hsp70 interacting protein); K09560 suppressor
of tumorigenicity protein 13
Length=379
Score = 124 bits (312), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 74/183 (40%), Positives = 112/183 (61%), Gaps = 3/183 (1%)
Query 11 DEEVVVPESTPAPPLAPAEERELTDEDYEKLAKAKEAATEAAEAGDLNRAVESYTEALLV 70
D E VVP P E E+T+E ++ + K A A G+L +A+E +TEA+ +
Sbjct 82 DNEGVVPGDDDEPQEMGDESVEVTEEMMDQANEKKVEAINALGEGELEKAIELFTEAIKL 141
Query 71 GNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHS 130
A+LY +RA V +K ++ AAIRDCD A+ +NPD+A+ Y+ RG A+R LGHW+ +
Sbjct 142 NPRIAILYAKRASVYVKLQKPNAAIRDCDRAIAINPDSAQPYKWRGKAHRLLGHWEDSAH 201
Query 131 DIEMGQKIDYDENIWDIQKLVEQKYKIIEEHERAAQRKKEEK---ERKERERRAREQRAN 187
D+ + K+DYDE+ + K V+ + I EH R +RK+EE+ ERKER ++A+E+
Sbjct 202 DLAIACKLDYDEDASTLLKEVQPRANKIAEHRRKYERKREEREINERKERLKKAKEENER 261
Query 188 AQR 190
AQR
Sbjct 262 AQR 264
> ath:AT3G17880 ATTDX; ATTDX (TETRATICOPEPTIDE DOMAIN-CONTAINING
THIOREDOXIN); oxidoreductase, acting on sulfur group of donors,
disulfide as acceptor / protein binding
Length=373
Score = 124 bits (311), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 73/178 (41%), Positives = 108/178 (60%), Gaps = 3/178 (1%)
Query 9 LNDEEVVVPESTPAPPLAPAEERELTDEDYEKLAKAKEAATEAAEAGDLNRAVESYTEAL 68
L++ +VV P++ P P+ E+TDE+ + K A EA G + A+E T+A+
Sbjct 74 LDNSDVVEPDNEPPQPMGDPTA-EVTDENRDDAQSEKSKAMEAISDGRFDEAIEHLTKAV 132
Query 69 LVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQA 128
++ +A+LY RA V LK K+ AAIRD + AL+ N D+A+ Y+ RG A LG W++A
Sbjct 133 MLNPTSAILYATRASVFLKVKKPNAAIRDANVALQFNSDSAKGYKSRGMAKAMLGQWEEA 192
Query 129 HSDIEMGQKIDYDENIWDIQKLVEQKYKIIEEHERAAQRKKEEKE--RKERERRAREQ 184
+D+ + K+DYDE I + K VE K IEEH R QR ++EKE R ERERR +++
Sbjct 193 AADLHVASKLDYDEEIGTMLKKVEPNAKRIEEHRRKYQRLRKEKELQRAERERRKQQE 250
> ath:AT4G22670 AtHip1; AtHip1 (Arabidopsis thaliana Hsp70-interacting
protein 1); binding
Length=441
Score = 122 bits (305), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 70/168 (41%), Positives = 101/168 (60%), Gaps = 0/168 (0%)
Query 32 ELTDEDYEKLAKAKEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRY 91
E+TDE+ E +AK A EA G+ + A+E T A+ + +A++Y RA V +K K+
Sbjct 114 EVTDENREAAQEAKGKAMEALSEGNFDEAIEHLTRAITLNPTSAIMYGNRASVYIKLKKP 173
Query 92 AAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKIDYDENIWDIQKLV 151
AAIRD + AL++NPD+A+ Y+ RG A LG W +A D+ + IDYDE I + K V
Sbjct 174 NAAIRDANAALEINPDSAKGYKSRGMARAMLGEWAEAAKDLHLASTIDYDEEISAVLKKV 233
Query 152 EQKYKIIEEHERAAQRKKEEKERKERERRAREQRANAQRAYEEQKKRE 199
E +EEH R R ++E+E K+ ER +RA AQ AY++ KK E
Sbjct 234 EPNAHKLEEHRRKYDRLRKEREDKKAERDRLRRRAEAQAAYDKAKKEE 281
> xla:446492 MGC79131 protein; K09560 suppressor of tumorigenicity
protein 13
Length=376
Score = 121 bits (304), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 71/183 (38%), Positives = 112/183 (61%), Gaps = 3/183 (1%)
Query 11 DEEVVVPESTPAPPLAPAEERELTDEDYEKLAKAKEAATEAAEAGDLNRAVESYTEALLV 70
D E V+P P E E+T+E ++ + K A A G+L +++E +TEA+ +
Sbjct 82 DNEGVIPGDDDEPQEMGDESAEVTEEMMDQANEKKVEAINALGEGELQKSIELFTEAIKL 141
Query 71 GNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHS 130
A+LY +RA V ++ ++ AAIRDCD A+ +NPD+A+ Y+ RG A+R LGHW+ +
Sbjct 142 NPRIAILYAKRASVYVQLQKPNAAIRDCDRAIAINPDSAQPYKWRGKAHRLLGHWEDSAH 201
Query 131 DIEMGQKIDYDENIWDIQKLVEQKYKIIEEHERAAQRKKEEKE---RKERERRAREQRAN 187
D+ + K+DYDE+ + K V+ + I EH R +RK+EEKE +KER ++A+E+
Sbjct 202 DLAIACKLDYDEDASAMLKEVQPRANKIAEHRRKHERKREEKEINDKKERLKKAKEENER 261
Query 188 AQR 190
AQR
Sbjct 262 AQR 264
> cel:T12D8.8 hypothetical protein; K09560 suppressor of tumorigenicity
protein 13
Length=422
Score = 119 bits (299), Expect = 5e-27, Method: Compositional matrix adjust.
Identities = 74/191 (38%), Positives = 117/191 (61%), Gaps = 1/191 (0%)
Query 9 LNDEEVVVPESTPAPPLAPAEERELTDEDYEKLAKAKEAATEAAEAGDLNRAVESYTEAL 68
+++E V+ PE A P+ + +E T+++ EK ++ + A EA GD + A+ +T A+
Sbjct 84 IDNEGVIEPEEAVALPMGDSA-KEATEDEIEKASEERGKAQEAFSNGDFDTALTHFTAAI 142
Query 69 LVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQA 128
+A+L+ +RA+VLLK KR AAI DCD+A+ +NPD+A+ Y+ RG ANR LG W +A
Sbjct 143 EANPGSAMLHAKRANVLLKLKRPVAAIADCDKAISINPDSAQGYKFRGRANRLLGKWVEA 202
Query 129 HSDIEMGQKIDYDENIWDIQKLVEQKYKIIEEHERAAQRKKEEKERKERERRAREQRANA 188
+D+ K+DYDE + K VE I+E+ RA +R+K + E ER R R +
Sbjct 203 KTDLATACKLDYDEAANEWLKEVEPNAHKIQEYNRAVERQKADIELAERRERVRRAQEAN 262
Query 189 QRAYEEQKKRE 199
++A EE+ KR+
Sbjct 263 KKAAEEEAKRQ 273
> hsa:6767 ST13, AAG2, FAM10A1, FAM10A4, FLJ27260, HIP, HOP, HSPABP,
HSPABP1, MGC129952, P48, PRO0786, SNC6; suppression of
tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein);
K09560 suppressor of tumorigenicity protein 13
Length=369
Score = 115 bits (289), Expect = 7e-26, Method: Compositional matrix adjust.
Identities = 69/192 (35%), Positives = 109/192 (56%), Gaps = 16/192 (8%)
Query 11 DEEVVVPESTPAPPLAPAEERELTDEDYEKLAKAKEAATEAAEAGDLNRAVESYTEALLV 70
D+E V+ T AP E E+T+E ++ K AA EA G+L +A++ +T+A+ +
Sbjct 84 DKEGVIEPDTDAPQEMGDENAEITEEMMDQANDKKVAAIEALNDGELQKAIDLFTDAIKL 143
Query 71 GNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHS 130
A+LY +RA V +K ++ AAIRDCD A+++NPD+A+ Y+ RG A+R LGHW++A
Sbjct 144 NPRLAILYAKRASVFVKLQKPNAAIRDCDRAIEINPDSAQPYKWRGKAHRLLGHWEEAAH 203
Query 131 DIEMGQKIDYDENIWDIQKLVEQKYKIIEEH----------------ERAAQRKKEEKER 174
D+ + K+DYDE+ + K V+ + + I EH ++ +EE ER
Sbjct 204 DLALACKLDYDEDASAMLKEVQPRAQKIAEHRRKYERKREEREIKERIERVKKAREEHER 263
Query 175 KERERRAREQRA 186
+RE AR Q
Sbjct 264 AQREEEARRQSG 275
> mmu:70356 St13, 1110007I03Rik, 3110002K08Rik, AW555194, HIP,
HOP, HSPABP, HSPABP1, PRO0786, SNC6, p48; suppression of tumorigenicity
13; K09560 suppressor of tumorigenicity protein
13
Length=371
Score = 115 bits (287), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 68/193 (35%), Positives = 110/193 (56%), Gaps = 16/193 (8%)
Query 11 DEEVVVPESTPAPPLAPAEERELTDEDYEKLAKAKEAATEAAEAGDLNRAVESYTEALLV 70
D E V+ T AP E E+T+E ++ + K AA EA G+L +A++ +T+A+ +
Sbjct 83 DNEGVIEPDTDAPQEMGDENAEITEEMMDEANEKKGAAIEALNDGELQKAIDLFTDAIKL 142
Query 71 GNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHS 130
A+LY +RA V +K ++ AAIRDCD A+++NPD+A+ Y+ RG A+R LGHW++A
Sbjct 143 NPRLAILYAKRASVFVKLQKPNAAIRDCDRAIEINPDSAQPYKWRGKAHRLLGHWEEAAH 202
Query 131 DIEMGQKIDYDENIWDIQKLVEQKYKIIEEH----------------ERAAQRKKEEKER 174
D+ + K+DYDE+ + + V+ + + I EH ++ +EE ER
Sbjct 203 DLALACKLDYDEDASAMLREVQPRAQKIAEHRRKYERKREEREIKERIERVKKAREEHER 262
Query 175 KERERRAREQRAN 187
+RE AR Q +
Sbjct 263 AQREEEARRQSGS 275
> dre:564225 st13, MGC73267, MGC77089, wu:fd15g02, zgc:73267;
suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting
protein); K09560 suppressor of tumorigenicity protein
13
Length=362
Score = 110 bits (276), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 65/191 (34%), Positives = 109/191 (57%), Gaps = 16/191 (8%)
Query 10 NDEEVVVPESTPAPPLAPAEERELTDEDYEKLAKAKEAATEAAEAGDLNRAVESYTEALL 69
++E V+ P++ + E E+T+E ++ + K A +A GDL +A++ +TEA+
Sbjct 86 DNEGVIEPDTDDPQEMGDFENLEVTEEMMDQANEKKTEAIDALGDGDLQKALDLFTEAIK 145
Query 70 VGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAH 129
+ A+LY +RA V +K ++ AAIRDCD A+ +NPD+A+ Y+ RG A++ LGHW+++
Sbjct 146 LNPKLAILYAKRASVYVKMQKPNAAIRDCDRAISINPDSAQPYKWRGKAHKLLGHWEESA 205
Query 130 SDIEMGQKIDYDENIWDIQKLVEQKYKIIEEH----------------ERAAQRKKEEKE 173
D+ M K+DYDE + K V+ K I +H + ++ +EE E
Sbjct 206 RDLAMACKLDYDEEASAMLKEVQPKANKIIDHRRKYERKREEREIRARQERVKKAREEHE 265
Query 174 RKERERRAREQ 184
R +RE AR+Q
Sbjct 266 RAQREEEARQQ 276
> hsa:10953 TOMM34, HTOM34P, TOM34, URCC3; translocase of outer
mitochondrial membrane 34
Length=309
Score = 65.5 bits (158), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 36/96 (37%), Positives = 55/96 (57%), Gaps = 0/96 (0%)
Query 37 DYEKLAKAKEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIR 96
D EK KE E + G+ +A+E Y+E+LL N + Y+ RA L K+Y A++
Sbjct 189 DVEKARVLKEEGNELVKKGNHKKAIEKYSESLLCSNLESATYSNRALCYLVLKQYTEAVK 248
Query 97 DCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSDI 132
DC EALKL+ N +A+ R A++ L +K + +DI
Sbjct 249 DCTEALKLDGKNVKAFYRRAQAHKALKDYKSSFADI 284
Score = 33.1 bits (74), Expect = 0.68, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 31/65 (47%), Gaps = 0/65 (0%)
Query 75 ALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEM 134
++LY+ RA LK I+DC AL L P + + R +A L + A+ D +
Sbjct 51 SVLYSNRAACHLKDGNCRDCIKDCTSALALVPFSIKPLLRRASAYEALEKYPMAYVDYKT 110
Query 135 GQKID 139
+ID
Sbjct 111 VLQID 115
> dre:768178 zgc:153288
Length=591
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 53/88 (60%), Gaps = 1/88 (1%)
Query 45 KEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKL 104
K+ EA + D A++ Y +L + + +A ++ RA L++ +++ AA+ DCD L+L
Sbjct 198 KDKGNEAYRSRDYEEALDYYCRSLSLAS-SAAVFNNRAQTLIRLQQWPAALSDCDAVLQL 256
Query 105 NPDNARAYRIRGTANRYLGHWKQAHSDI 132
P N +A R T +++LGH +++H D+
Sbjct 257 EPHNIKALLRRATVHKHLGHQQESHDDL 284
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 31/102 (30%), Positives = 57/102 (55%), Gaps = 2/102 (1%)
Query 62 ESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRY 121
E +T++L + T YT RA +K +R+ A +DCD AL++ P N +A+ R AN+
Sbjct 471 ERHTQSLQLDTHTCAAYTNRALCYIKLERFTEARQDCDSALQIEPTNKKAFYRRALANKG 530
Query 122 LGHWKQAHSDIEMGQKIDYDENIWDIQKLVEQKYKIIEEHER 163
L + SD++ Q + D ++ + Q+L+ + ++E+ R
Sbjct 531 LKDYLSCRSDLQ--QVLRLDASVTEAQRLLMELTHLMEDRRR 570
> hsa:6674 SPAG1, FLJ32920, HSD-3.8, SP75, TPIS; sperm associated
antigen 1
Length=926
Score = 57.0 bits (136), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 46/174 (26%), Positives = 84/174 (48%), Gaps = 18/174 (10%)
Query 16 VPESTPAPPLAPAEE--------------RELTDEDYEKLAKAKEAATEAAEAGDLNRAV 61
VP S P PA+E + +TDE + KE + + A+
Sbjct 586 VPASVPLQAWHPAKEMISKQAGDSSSHRQQGITDE--KTFKALKEEGNQCVNDKNYKDAL 643
Query 62 ESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRY 121
Y+E L + N +YT RA LK ++ A +DCD+AL+L N +A+ R A++
Sbjct 644 SKYSECLKINNKECAIYTNRALCYLKLCQFEEAKQDCDQALQLADGNVKAFYRRALAHKG 703
Query 122 LGHWKQAHSDIEMGQKIDYDENIWDIQKLVEQKYKIIEEHERAAQRKKEEKERK 175
L ++++ S I++ + I D +I + + +E+ +++ ++ A KE++ RK
Sbjct 704 LKNYQK--SLIDLNKVILLDPSIIEAKMELEEVTRLLNLKDKTAPFNKEKERRK 755
Score = 50.4 bits (119), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 60/113 (53%), Gaps = 2/113 (1%)
Query 33 LTDEDYEKLA-KAKEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRY 91
LT+++ + LA + KE EA +GD AV YT ++ PT + Y RA +K + +
Sbjct 200 LTEKEKDFLATREKEKGNEAFNSGDYEEAVMYYTRSI-SALPTVVAYNNRAQAEIKLQNW 258
Query 92 AAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKIDYDENI 144
+A +DC++ L+L P N +A R T ++ ++A D+ ++ D ++
Sbjct 259 NSAFQDCEKVLELEPGNVKALLRRATTYKHQNKLREATEDLSKVLDVEPDNDL 311
Score = 43.5 bits (101), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 28/106 (26%), Positives = 48/106 (45%), Gaps = 8/106 (7%)
Query 42 AKAKEAATEAAEAGDLNRAVESYTEALLVGNPT--------ALLYTRRADVLLKQKRYAA 93
A K E +G A Y+ A+ + P ++LY+ RA LK+ +
Sbjct 446 AGLKSQGNELFRSGQFAEAAGKYSAAIALLEPAGSEIADDLSILYSNRAACYLKEGNCSG 505
Query 94 AIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKID 139
I+DC+ AL+L+P + + R A L + +A+ D + +ID
Sbjct 506 CIQDCNRALELHPFSMKPLLRRAMAYETLEQYGKAYVDYKTVLQID 551
> ath:AT1G56440 serine/threonine protein phosphatase-related
Length=476
Score = 57.0 bits (136), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 44/132 (33%), Positives = 67/132 (50%), Gaps = 8/132 (6%)
Query 45 KEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKL 104
KE E + N A++ Y+ ++ + +P A+ Y RA LK KRY A DC EAL L
Sbjct 88 KEQGNEFFKQKKFNEAIDCYSRSIAL-SPNAVTYANRAMAYLKIKRYREAEVDCTEALNL 146
Query 105 NPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKI-----DYDENIWDIQKLVEQKYKIIE 159
+ +AY R TA + LG K+A D E ++ + + DI+ L+E+ +IIE
Sbjct 147 DDRYIKAYSRRATARKELGMIKEAKEDAEFALRLEPESQELKKQYADIKSLLEK--EIIE 204
Query 160 EHERAAQRKKEE 171
+ A Q +E
Sbjct 205 KATGAMQSTAQE 216
> dre:564953 spag1, MGC162178, cb1089, wu:fj78g10; sperm associated
antigen 1
Length=386
Score = 56.6 bits (135), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 33/116 (28%), Positives = 60/116 (51%), Gaps = 2/116 (1%)
Query 45 KEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKL 104
K+ E + A E Y+E L + +YT RA LK +R+A A +DCD AL++
Sbjct 265 KQEGNELVKNSQFQGASEKYSECLAIKPNECAIYTNRALCFLKLERFAEAKQDCDSALQM 324
Query 105 NPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKIDYDENIWDIQKLVEQKYKIIEE 160
P N +A+ R A++ L + A +D++ + + D N+ + ++ +E ++ E
Sbjct 325 EPKNKKAFYRRALAHKGLKDYLSASTDLQ--EVLQLDPNVQEAEQELEMVTNLLRE 378
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 40/150 (26%), Positives = 62/150 (41%), Gaps = 26/150 (17%)
Query 16 VPESTPAP------------PLAPAEERELTDEDYE------KLAKAKEAATEAAEAGDL 57
VP S+PA P P E + D LA+ K + G
Sbjct 41 VPGSSPAANGSLPAGKSQDEPQGPGSAGESCNLDAPCGALPPPLARLKNQGNMLFKNGQF 100
Query 58 NRAVESYTEAL------LVGNP--TALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNA 109
A+E YT+A+ + +P +LY+ RA LK A I+DC AL+L+P +
Sbjct 101 GDALEKYTQAIDGCIEAGIDSPEDLCVLYSNRAACFLKDGNSADCIQDCTRALELHPFSL 160
Query 110 RAYRIRGTANRYLGHWKQAHSDIEMGQKID 139
+ R A L +++A+ D + +ID
Sbjct 161 KPLLRRAMAYESLERYRKAYVDYKTVLQID 190
> xla:414472 rpap3, MGC81126; RNA polymerase II associated protein
3
Length=660
Score = 55.8 bits (133), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 31/80 (38%), Positives = 44/80 (55%), Gaps = 0/80 (0%)
Query 60 AVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTAN 119
A+E Y++ + N ALL RA LK ++Y A DC A+ L+ +A+ RGTA+
Sbjct 303 AIECYSQGMEADNTNALLPANRAMAYLKIQKYKEAEADCTLAISLDASYCKAFARRGTAS 362
Query 120 RYLGHWKQAHSDIEMGQKID 139
LG K+A D EM K+D
Sbjct 363 IMLGKQKEAKEDFEMVLKLD 382
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 49/163 (30%), Positives = 81/163 (49%), Gaps = 11/163 (6%)
Query 37 DYEKLAKAKEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIR 96
D EK KE ++G + A+E YT + A+L T RA + K++A A
Sbjct 128 DTEKALSEKEKGNNYFKSGKYDEAIECYTRGMDADPYNAILPTNRASAFFRLKKFAVAES 187
Query 97 DCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKIDYDENIW----DIQKLVE 152
DC+ A+ LN D A+AY RG A L + + A D E + ++ D N + +++K+ +
Sbjct 188 DCNLAIALNRDYAKAYARRGAARLALKNLQGAKEDYE--KVLELDANNFEAKNELRKINQ 245
Query 153 QKYKIIEE-HERAAQRKK----EEKERKERERRAREQRANAQR 190
+ Y + E A K E+E+K+ E + R+Q+A Q+
Sbjct 246 ELYSSASDVQENMATEAKITVENEEEKKQIEIQQRKQQAIMQK 288
> cel:Y39B6A.2 pph-5; Protein PHosphatase family member (pph-5);
K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=496
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 38/117 (32%), Positives = 63/117 (53%), Gaps = 2/117 (1%)
Query 22 APPLAPAEERELTDEDYEKLAKAKEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRR 81
A L EE+ DE EK K+ A + + + A + Y+ A+ + +PTA+LY R
Sbjct 11 ATVLESIEEKSYEDEK-EKAGMIKDEANQFFKDQVYDVAADLYSVAIEI-HPTAVLYGNR 68
Query 82 ADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKI 138
A LK++ Y +A+ D D A+ ++P + + R TAN LG +K+A +D + K+
Sbjct 69 AQAYLKKELYGSALEDADNAIAIDPSYVKGFYRRATANMALGRFKKALTDYQAVVKV 125
> ath:AT5G09420 ATTOC64-V (ARABIDOPSIS THALIANA TRANSLOCON AT
THE OUTER MEMBRANE OF CHLOROPLASTS 64-V); amidase/ binding /
carbon-nitrogen ligase, with glutamine as amido-N-donor
Length=603
Score = 55.1 bits (131), Expect = 1e-07, Method: Composition-based stats.
Identities = 36/110 (32%), Positives = 54/110 (49%), Gaps = 3/110 (2%)
Query 22 APPLAPAEERELTDEDYEKLAKAKEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRR 81
A LAP + T+ + E KE A + N+AV YTEA+ + A Y R
Sbjct 472 ASNLAPVSD---TNGNMEASEVMKEKGNAAYKGKQWNKAVNFYTEAIKLNGANATYYCNR 528
Query 82 ADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSD 131
A L+ + A +DC +A+ ++ N +AY RGTA L +K+A +D
Sbjct 529 AAAFLELCCFQQAEQDCTKAMLIDKKNVKAYLRRGTARESLVRYKEAAAD 578
> hsa:79657 RPAP3, FLJ21908; RNA polymerase II associated protein
3
Length=631
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 29/79 (36%), Positives = 42/79 (53%), Gaps = 0/79 (0%)
Query 55 GDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRI 114
G RA+E YT + ALL RA LK ++Y A +DC +A+ L+ ++A+
Sbjct 296 GKYERAIECYTRGIAADGANALLPANRAMAYLKIQKYEEAEKDCTQAILLDGSYSKAFAR 355
Query 115 RGTANRYLGHWKQAHSDIE 133
RGTA +LG +A D E
Sbjct 356 RGTARTFLGKLNEAKQDFE 374
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 29/97 (29%), Positives = 46/97 (47%), Gaps = 0/97 (0%)
Query 37 DYEKLAKAKEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIR 96
D +K KE + + G + A++ YT+ + +L T RA + K++A A
Sbjct 129 DSQKALVLKEKGNKYFKQGKYDEAIDCYTKGMDADPYNPVLPTNRASAYFRLKKFAVAES 188
Query 97 DCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSDIE 133
DC+ A+ LN +AY RG A L ++A D E
Sbjct 189 DCNLAVALNRSYTKAYSRRGAARFALQKLEEAKKDYE 225
> mmu:26942 Spag1, tpis; sperm associated antigen 1
Length=901
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 36/113 (31%), Positives = 64/113 (56%), Gaps = 2/113 (1%)
Query 33 LTDEDYEKLA-KAKEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRY 91
LT+++ LA + K EA +GD AV YT +L PTA+ Y RA +K +R+
Sbjct 204 LTEKEKSFLANREKGKGNEAFYSGDYEEAVMYYTRSL-SALPTAIAYNNRAQAEIKLQRW 262
Query 92 AAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKIDYDENI 144
++A+ DC++AL+L+P N +A R T ++ ++A D+ +++ D ++
Sbjct 263 SSALEDCEKALELDPGNVKALLRRATTYKHQNKLQEAVDDLRKVLQVEPDNDL 315
Score = 39.7 bits (91), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 44/100 (44%), Gaps = 8/100 (8%)
Query 48 ATEAAEAGDLNRAVESYTEALLVGNPT--------ALLYTRRADVLLKQKRYAAAIRDCD 99
E G A Y+ A+ PT ++LY+ RA LK+ I+DC+
Sbjct 437 GNELFRGGQFAEAAAQYSVAIAQLEPTGSANADELSILYSNRAACYLKEGNCRDCIQDCN 496
Query 100 EALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKID 139
AL+L+P + + R A L ++ A+ D + +ID
Sbjct 497 RALELHPFSVKPLLRRAMAYETLEQYRNAYVDYKTVLQID 536
Score = 38.9 bits (89), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 31/118 (26%), Positives = 57/118 (48%), Gaps = 9/118 (7%)
Query 16 VPESTPAPP-LAPAEERELTDEDYEKLAKA-KEAATEAAEAGDLNRAVESYTEALLVGNP 73
P+ P P P+ +TDE K+ +A KE + + + A+ Y E L + +
Sbjct 586 TPDQDPCPNNCMPS----ITDE---KMFQALKEEGNQLVKDKNYKDAISKYNECLKINSK 638
Query 74 TALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSD 131
+YT RA LK ++ A DC++AL+++ +N +A A + L + +++ D
Sbjct 639 ACAIYTNRALCYLKLGQFEEAKLDCEQALQIDGENVKASHRLALAQKGLENCRESGVD 696
Score = 33.5 bits (75), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 29/60 (48%), Gaps = 1/60 (1%)
Query 85 LLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKIDYDENI 144
L+K K Y AI +E LK+N Y R LG +++A D E +ID EN+
Sbjct 616 LVKDKNYKDAISKYNECLKINSKACAIYTNRALCYLKLGQFEEAKLDCEQALQID-GENV 674
> mmu:71919 Rpap3, 2310042P20Rik, D15Ertd682e; RNA polymerase
II associated protein 3
Length=660
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/79 (37%), Positives = 42/79 (53%), Gaps = 0/79 (0%)
Query 55 GDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRI 114
G +A+E YT + ALL RA LK +RY A RDC +A+ L+ ++A+
Sbjct 298 GKYEQAIECYTRGIAADRTNALLPANRAMAYLKIQRYEEAERDCTQAIVLDGSYSKAFAR 357
Query 115 RGTANRYLGHWKQAHSDIE 133
RGTA +LG +A D E
Sbjct 358 RGTARTFLGKINEAKQDFE 376
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 29/97 (29%), Positives = 45/97 (46%), Gaps = 0/97 (0%)
Query 37 DYEKLAKAKEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIR 96
D +K KE + + G + A+E YT+ + +L T RA + K++A A
Sbjct 130 DSQKALVLKEKGNKYFKQGKYDEAIECYTKGMDADPYNPVLPTNRASAYFRLKKFAVAES 189
Query 97 DCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSDIE 133
DC+ A+ L+ +AY RG A L + A D E
Sbjct 190 DCNLAIALSRTYTKAYARRGAARFALQKLEDARKDYE 226
> sce:YGR123C PPT1; Ppt1p (EC:3.1.3.16); K04460 protein phosphatase
5 [EC:3.1.3.16]
Length=513
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 26/76 (34%), Positives = 46/76 (60%), Gaps = 0/76 (0%)
Query 59 RAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTA 118
+A+E YTEA+ + + ++ ++ RA K + +A+ DCDEA+KL+P N +AY R +
Sbjct 30 KAIEKYTEAIDLDSTQSIYFSNRAFAHFKVDNFQSALNDCDEAIKLDPKNIKAYHRRALS 89
Query 119 NRYLGHWKQAHSDIEM 134
L +K+A D+ +
Sbjct 90 CMALLEFKKARKDLNV 105
> dre:493606 stip1, zgc:92133; stress-induced-phosphoprotein 1
(Hsp70/Hsp90-organizing protein); K09553 stress-induced-phosphoprotein
1
Length=542
Score = 53.5 bits (127), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 52/97 (53%), Gaps = 0/97 (0%)
Query 39 EKLAKAKEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDC 98
EK+++ K+ +A AG+L A+ YTEAL + +L++ R+ K+ Y A++D
Sbjct 2 EKVSQLKDQGNKALSAGNLEEAIRCYTEALTLDPSNHVLFSNRSAAYAKKGDYDNALKDA 61
Query 99 DEALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEMG 135
+ +K+ PD + Y + A +LG + A + + G
Sbjct 62 CQTIKIKPDWGKGYSRKAAALEFLGRLEDAKATYQEG 98
Score = 38.1 bits (87), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 37/74 (50%), Gaps = 0/74 (0%)
Query 45 KEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKL 104
K +A + GD A++ Y+EA+ A L++ RA K + A++DC+E + L
Sbjct 363 KNKGNDAFQKGDYPLAMKHYSEAIKRNPYDAKLFSNRAACYTKLLEFQLALKDCEECINL 422
Query 105 NPDNARAYRIRGTA 118
+ + Y +G A
Sbjct 423 DSTFIKGYTRKGAA 436
> mmu:67145 Tomm34, 2610100K07Rik, TOM34, Tomm34a, Tomm34b; translocase
of outer mitochondrial membrane 34
Length=309
Score = 53.5 bits (127), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 31/90 (34%), Positives = 50/90 (55%), Gaps = 0/90 (0%)
Query 37 DYEKLAKAKEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIR 96
D E+ KE + + G+ +A+E Y+E+LL + + Y+ RA L K+Y A++
Sbjct 189 DVERAKALKEEGNDLVKKGNHKKAIEKYSESLLCSSLESATYSNRALCHLVLKQYKEAVK 248
Query 97 DCDEALKLNPDNARAYRIRGTANRYLGHWK 126
DC EALKL+ N +A+ R A + L +K
Sbjct 249 DCTEALKLDGKNVKAFYRRAQAYKALKDYK 278
Score = 34.3 bits (77), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 31/65 (47%), Gaps = 0/65 (0%)
Query 75 ALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEM 134
++LY+ RA LK I+DC AL L P + + R +A L + A+ D +
Sbjct 51 SVLYSNRAACYLKDGNCTDCIKDCTSALALVPFSIKPLLRRASAYEALEKYALAYVDYKT 110
Query 135 GQKID 139
+ID
Sbjct 111 VLQID 115
> sce:YOR027W STI1; Hsp90 cochaperone, interacts with the Ssa
group of the cytosolic Hsp70 chaperones; activates the ATPase
activity of Ssa1p; homolog of mammalian Hop protein; K09553
stress-induced-phosphoprotein 1
Length=589
Score = 52.8 bits (125), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 28/96 (29%), Positives = 53/96 (55%), Gaps = 1/96 (1%)
Query 45 KEAATEAAEAGDLNRAVESYTEALLVG-NPTALLYTRRADVLLKQKRYAAAIRDCDEALK 103
K+ A A D ++A+E +T+A+ V P +LY+ R+ K+++ A+ D +E +K
Sbjct 9 KQQGNAAFTAKDYDKAIELFTKAIEVSETPNHVLYSNRSACYTSLKKFSDALNDANECVK 68
Query 104 LNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKID 139
+NP ++ Y G A+ LG +A S+ + ++D
Sbjct 69 INPSWSKGYNRLGAAHLGLGDLDEAESNYKKALELD 104
Score = 40.4 bits (93), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 34/116 (29%), Positives = 56/116 (48%), Gaps = 3/116 (2%)
Query 43 KAKEAATEAAE---AGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCD 99
KA+EA E E D AV++YTE + A Y+ RA L K + AI DC+
Sbjct 395 KAEEARLEGKEYFTKSDWPNAVKAYTEMIKRAPEDARGYSNRAAALAKLMSFPEAIADCN 454
Query 100 EALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKIDYDENIWDIQKLVEQKY 155
+A++ +P+ RAY + TA + + A ++ + D + N + ++Q Y
Sbjct 455 KAIEKDPNFVRAYIRKATAQIAVKEYASALETLDAARTKDAEVNNGSSAREIDQLY 510
> xla:414520 hypothetical protein MGC81394
Length=312
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 34/102 (33%), Positives = 52/102 (50%), Gaps = 7/102 (6%)
Query 34 TDEDYEKLAKAKEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAA 93
+DED + K E + + AV YT+AL + A+ Y RA K YA
Sbjct 81 SDEDVAEAESLKTEGNEQMKVENFESAVTYYTKALELNPRNAVYYCNRAAAYSKLGNYAG 140
Query 94 AIRDCDEALKLNPDNARAYRIRGTA----NRY---LGHWKQA 128
A+RDC+EA+ ++P ++AY G A N++ +G +KQA
Sbjct 141 AVRDCEEAISIDPSYSKAYGRMGLALSSLNKHAESVGFYKQA 182
> dre:323361 tomm34, wu:fb96b08, zgc:56645; translocase of outer
mitochondrial membrane 34
Length=305
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 41/114 (35%), Positives = 60/114 (52%), Gaps = 4/114 (3%)
Query 45 KEAATEAAEAGDLNRAVESYTEALLVGNPT-ALLYTRRADVLLKQKRYAAAIRDCDEALK 103
KE + G+ +A+E YT++L +PT YT RA L K Y AIRDC+EAL+
Sbjct 194 KEEGNALVKKGEHKKAMEKYTQSL-AQDPTEVTTYTNRALCYLALKMYKDAIRDCEEALR 252
Query 104 LNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKIDYDENIWDIQKLVEQKYKI 157
L+ N +A R A + L + K D+ KI D N +QKL+++ K+
Sbjct 253 LDSANIKALYRRAQAYKELKNKKSCIEDLNSVLKI--DPNNTAVQKLLQEVQKM 304
Score = 42.0 bits (97), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 30/105 (28%), Positives = 48/105 (45%), Gaps = 8/105 (7%)
Query 45 KEAATEAAEAGDLNRAVESYTEALLVGNPTA--------LLYTRRADVLLKQKRYAAAIR 96
K+A E +AG AV Y++A+ + +LY+ RA LK I+
Sbjct 14 KQAGNECFKAGQYGEAVTLYSQAIQQLEKSGQKKTEDLGILYSNRAASYLKDGNCNECIK 73
Query 97 DCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKIDYD 141
DC +L L P +A R A L ++QA+ D + +ID++
Sbjct 74 DCTASLDLVPFGFKALLRRAAAFEALERYRQAYVDYKTVLQIDWN 118
> xla:496358 sgta; small glutamine-rich tetratricopeptide repeat
(TPR)-containing, alpha
Length=302
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 32/102 (31%), Positives = 53/102 (51%), Gaps = 7/102 (6%)
Query 34 TDEDYEKLAKAKEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAA 93
+DED + + K E + + A+ YT+AL + A+ Y RA K YA
Sbjct 69 SDEDLAEAERLKTEGNEQMKVENFESAISYYTKALELNPANAVYYCNRAAAYSKLGNYAG 128
Query 94 AIRDCDEALKLNPDNARAYRIRGTA----NRY---LGHWKQA 128
A+RDC+ A+ ++P+ ++AY G A N++ +G +KQA
Sbjct 129 AVRDCEAAITIDPNYSKAYGRMGLALSSLNKHAEAVGFYKQA 170
> bbo:BBOV_III002230 17.m07215; tetratricopeptide repeat domain
containing protein; K09553 stress-induced-phosphoprotein 1
Length=546
Score = 50.8 bits (120), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 29/99 (29%), Positives = 50/99 (50%), Gaps = 0/99 (0%)
Query 41 LAKAKEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDE 100
+A K+ EA +AG AV+ +T A+ +LY+ R+ +R+ A+ D ++
Sbjct 1 MADHKQLGNEAFKAGRFLDAVQHFTAAIQANPSDGILYSNRSGAYASLQRFQEALDDANQ 60
Query 101 ALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKID 139
+ L PD + Y +G A LG ++A + + G KID
Sbjct 61 CVSLKPDWPKGYSRKGLALYKLGRLQEARTAYQEGLKID 99
Score = 40.0 bits (92), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 26/104 (25%), Positives = 54/104 (51%), Gaps = 6/104 (5%)
Query 72 NPTAL-LYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHS 130
NP+ + LYT RA L K Y +A+ DC++A++++P +A+ +G + L + +A
Sbjct 390 NPSDIKLYTNRAAALTKLGEYPSALADCNKAVEMDPTFVKAWARKGNLHVLLKEYSKALE 449
Query 131 DIEMGQKIDYDENIWDIQKLVEQKYKIIEEHERAAQRKKEEKER 174
+ G +D + Q+ + KY + + + +Q ++E+
Sbjct 450 AYDKGLALDPNN-----QECITGKYDCMAKIQAMSQSGTVDEEQ 488
> dre:563791 ttc12; tetratricopeptide repeat domain 12
Length=700
Score = 50.4 bits (119), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 32/84 (38%), Positives = 42/84 (50%), Gaps = 0/84 (0%)
Query 45 KEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKL 104
+E EA GD AV YTE L LYT RA +K KRY AI DC+ AL+
Sbjct 117 REQGNEAFTQGDYETAVRFYTEGLEQLRDMQALYTNRAQAFIKLKRYKEAISDCEWALRC 176
Query 105 NPDNARAYRIRGTANRYLGHWKQA 128
N +A+ GT++ L + Q+
Sbjct 177 NEKCIKAFIHMGTSHLALKDFTQS 200
> mmu:209683 Ttc28, 2310015L07Rik, 6030435N04, AI428795, AI851761,
BC002262, MGC7623; tetratricopeptide repeat domain 28
Length=2481
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/76 (35%), Positives = 42/76 (55%), Gaps = 0/76 (0%)
Query 48 ATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPD 107
+ +A GD + A+ Y EAL V +LY+ R+ +K ++Y A+ D +A LNP
Sbjct 59 SNQACHDGDFHTAIVLYNEALAVDPQNCILYSNRSAAYMKTQQYHKALDDAIKARLLNPK 118
Query 108 NARAYRIRGTANRYLG 123
+AY +G A +YLG
Sbjct 119 WPKAYFRQGVALQYLG 134
> tpv:TP03_0587 hypothetical protein; K09553 stress-induced-phosphoprotein
1
Length=540
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 43/73 (58%), Gaps = 1/73 (1%)
Query 72 NPT-ALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHS 130
NPT A LY+ RA LLK Y +A+ DC++AL+L+P +A+ +G + L + +A
Sbjct 385 NPTDAKLYSNRAAALLKLCEYPSALADCNKALELDPTFVKAWARKGNLHVLLKEYHKAMD 444
Query 131 DIEMGQKIDYDEN 143
+ G K+D + N
Sbjct 445 SYDKGLKVDPNNN 457
Score = 43.5 bits (101), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 28/99 (28%), Positives = 47/99 (47%), Gaps = 2/99 (2%)
Query 45 KEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKL 104
K +A +AG AVE +T+A+ + +LY+ R+ Y A+ D ++ + L
Sbjct 5 KNLGNDAFKAGRFMDAVEFFTKAIELNPDDHVLYSNRSGAYASMYMYNEALADANKCIDL 64
Query 105 NPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKIDYDEN 143
PD + Y +G LG+ ++A MG + YD N
Sbjct 65 KPDWPKGYSRKGLCEYKLGNPEKAKETYNMG--LAYDPN 101
> tgo:TGME49_047000 TPR domain-containing protein (EC:3.4.21.72)
Length=888
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 35/65 (53%), Gaps = 0/65 (0%)
Query 75 ALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEM 134
A+L + R L K + A + DC EA++ +P A+AY R TAN L W A +DI
Sbjct 744 AVLLSNRGACHLHGKCWEAVVADCTEAIQCDPSYAKAYLRRFTANEALTKWHDAAADINK 803
Query 135 GQKID 139
++D
Sbjct 804 AIELD 808
> xla:379955 stip1, MGC53256; stress-induced-phosphoprotein 1;
K09553 stress-induced-phosphoprotein 1
Length=543
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 27/91 (29%), Positives = 48/91 (52%), Gaps = 0/91 (0%)
Query 45 KEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKL 104
KE +A AG+L+ AV+ YTEA+ + +LY+ R+ K+K + A+ D + ++L
Sbjct 8 KEKGNKALSAGNLDEAVKCYTEAIKLDPKNHVLYSNRSAAYAKKKEFTKALEDGSKTVEL 67
Query 105 NPDNARAYRIRGTANRYLGHWKQAHSDIEMG 135
D + Y + A +L +++A E G
Sbjct 68 KADWGKGYSRKAAALEFLNRFEEAKKTYEEG 98
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/96 (26%), Positives = 47/96 (48%), Gaps = 0/96 (0%)
Query 44 AKEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALK 103
AK E+ + GD +A++ Y+EA+ A LY+ RA K + A++DC+E ++
Sbjct 363 AKNKGNESFQKGDYPQAMKHYSEAIKRNPNDAKLYSNRAACYTKLLEFLLAVKDCEECIR 422
Query 104 LNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKID 139
L P + Y + A + + +A + ++D
Sbjct 423 LEPSFIKGYTRKAAALEAMKDFTKAMDAYQKAMELD 458
Score = 30.8 bits (68), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 33/128 (25%), Positives = 62/128 (48%), Gaps = 6/128 (4%)
Query 43 KAKEAATEAAEAGDLNRAVESYTEALLVGNPTALLY-TRRADVLLKQKRYAAAIRDCDEA 101
K KE EA + D A++ Y +A + +P + Y T +A V + Y+ C++A
Sbjct 227 KEKELGNEAYKKKDFETALKHYGQAREL-DPANMTYITNQAAVYFEMGDYSKCRELCEKA 285
Query 102 LKLNPDNARAYRIRGTANRYLG--HWKQAHSD--IEMGQKIDYDENIWDIQKLVEQKYKI 157
+++ +N YR+ A +G ++K+ + I+ K + ++ K +Q KI
Sbjct 286 IEVGRENREDYRLIAKAYARIGNSYFKEEKNKEAIQFFNKSLAEHRTPEVLKKCQQAEKI 345
Query 158 IEEHERAA 165
++E ER A
Sbjct 346 LKEQERVA 353
> mmu:100048137 tetratricopeptide repeat protein 12-like
Length=544
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/84 (35%), Positives = 42/84 (50%), Gaps = 0/84 (0%)
Query 45 KEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKL 104
KE EA GD A+ Y+E L +LYT RA +K Y A+ DCD ALK
Sbjct 109 KEKGNEAFVRGDYETAIFFYSEGLGKLKDMKVLYTNRAQAFIKLGDYQKALVDCDWALKC 168
Query 105 NPDNARAYRIRGTANRYLGHWKQA 128
+ + +AY G A+ L ++ +A
Sbjct 169 DENCTKAYFHMGKAHVALKNYSKA 192
> ath:AT4G37460 SRFR1; SRFR1 (SUPPRESSOR OF RPS4-RLD 1); protein
complex scaffold
Length=1052
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 54/196 (27%), Positives = 83/196 (42%), Gaps = 12/196 (6%)
Query 12 EEVVVPESTPAPPLAPAEERELTDE--DYEKLAKAKEAATEA--------AEAGDLNRAV 61
EE PE+ A A +REL D+ K ++ AA+EA A G+ AV
Sbjct 326 EEPTYPEALIGRGTAYAFQRELESAIADFTKAIQSNPAASEAWKRRGQARAALGEYVEAV 385
Query 62 ESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRY 121
E T+AL+ + + R V K K + AA++D LK DN AY G A
Sbjct 386 EDLTKALVFEPNSPDVLHERGIVNFKSKDFTAAVKDLSICLKQEKDNKSAYTYLGLAFAS 445
Query 122 LGHWKQAHSDIEMGQKIDYDENIWDIQKLVEQKYKIIEEHERAAQRKKEEKERKERERRA 181
LG +K+A + I D N + + Q Y+ + +H +A + ++ + R +A
Sbjct 446 LGEYKKAEE--AHLKSIQLDSNYLEAWLHLAQFYQELADHCKALECIEQVLQVDNRVWKA 503
Query 182 REQRANAQRAYEEQKK 197
R E +K
Sbjct 504 YHLRGLVFHGLGEHRK 519
Score = 32.7 bits (73), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 37/72 (51%), Gaps = 0/72 (0%)
Query 66 EALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHW 125
++LL + L RA + + + I+DCD+AL L P +A+ ++G A LG
Sbjct 30 DSLLAKESSILDICNRAFCYNQLELHKHVIKDCDKALLLEPFAIQAFILKGRALLALGRK 89
Query 126 KQAHSDIEMGQK 137
++A +E G K
Sbjct 90 QEAVLVLEQGYK 101
> dre:431772 sgta, zgc:92462; small glutamine-rich tetratricopeptide
repeat (TPR)-containing, alpha
Length=306
Score = 47.8 bits (112), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 41/160 (25%), Positives = 72/160 (45%), Gaps = 21/160 (13%)
Query 2 DTFEPESLNDEEVVVPESTPAPPLAPAEERELTDEDYEKLAKAKEAATEAAEAGDLNRAV 61
+ F L ++ V +P++ P+P ED E+ + K + + + AV
Sbjct 60 EIFLNSLLKNDIVTLPKTFPSP------------EDIERAEQLKNEGNNHMKEENYSSAV 107
Query 62 ESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRY 121
+ YT+A+ + A+ Y RA K + Y A+ DC+ A+ ++P ++AY G A
Sbjct 108 DCYTKAIELDQRNAVYYCNRAAAHSKLENYTEAMGDCERAIAIDPSYSKAYGRMGLALTS 167
Query 122 LGHWKQAHSDIEMGQKID-----YDENIWDIQKLVEQKYK 156
+ + +A S +D Y N+ K+VEQK K
Sbjct 168 MSKYPEAISYFNKALVLDPENDTYKSNL----KIVEQKQK 203
> xla:398004 ppp5c, pp5; protein phosphatase 5, catalytic subunit
(EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=493
Score = 47.0 bits (110), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 36/115 (31%), Positives = 53/115 (46%), Gaps = 5/115 (4%)
Query 45 KEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKL 104
KE A E D + AV+ YT+A+ + TA+ Y R+ L+ + Y A+ D A++L
Sbjct 26 KEQANEYFRVKDYDHAVQYYTQAIDLSPDTAIYYGNRSLAYLRTECYGYALADASRAIQL 85
Query 105 NPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKI-----DYDENIWDIQKLVEQK 154
+ + Y R +N LG K A D E K+ D + KLV QK
Sbjct 86 DAKYIKGYYRRAASNMALGKLKAALKDYETVVKVRPHDKDAQMKFQECNKLVRQK 140
> ath:AT1G33400 tetratricopeptide repeat (TPR)-containing protein
Length=798
Score = 47.0 bits (110), Expect = 4e-05, Method: Composition-based stats.
Identities = 29/93 (31%), Positives = 48/93 (51%), Gaps = 9/93 (9%)
Query 56 DLNRAVESYTEALLVGNPTAL---------LYTRRADVLLKQKRYAAAIRDCDEALKLNP 106
D + A+ Y++AL V A+ L+ RA+VL ++RDC AL+++P
Sbjct 78 DFDEALRLYSKALRVAPLDAIDGDKSLLASLFLNRANVLHNLGLLKESLRDCHRALRIDP 137
Query 107 DNARAYRIRGTANRYLGHWKQAHSDIEMGQKID 139
A+A+ RG N LG++K A DI + ++
Sbjct 138 YYAKAWYRRGKLNTLLGNYKDAFRDITVSMSLE 170
> mmu:235330 Ttc12, E330017O07Rik; tetratricopeptide repeat domain
12
Length=704
Score = 46.6 bits (109), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 30/84 (35%), Positives = 42/84 (50%), Gaps = 0/84 (0%)
Query 45 KEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKL 104
KE EA GD A+ Y+E L +LYT RA +K Y A+ DCD ALK
Sbjct 109 KEKGNEAFVRGDYETAIFFYSEGLGKLKDMKVLYTNRAQAFIKLGDYQKALVDCDWALKC 168
Query 105 NPDNARAYRIRGTANRYLGHWKQA 128
+ + +AY G A+ L ++ +A
Sbjct 169 DENCTKAYFHMGKAHVALKNYSKA 192
> dre:393173 MGC56178; zgc:56178
Length=595
Score = 46.6 bits (109), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 36/94 (38%), Positives = 48/94 (51%), Gaps = 2/94 (2%)
Query 60 AVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTAN 119
AVESYT + ALL RA LK R+A A +DC AL L+P +A+ R TA
Sbjct 298 AVESYTRGMEADETNALLPANRAMAFLKLNRFAEAEQDCSAALALDPSYTKAFARRATAR 357
Query 120 RYLGHWKQAHSDIEMGQKID--YDENIWDIQKLV 151
LG + A D E K++ + I +I+KL
Sbjct 358 AALGKCRDARDDFEQVLKLEPGNKQAISEIEKLT 391
Score = 43.9 bits (102), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 28/95 (29%), Positives = 45/95 (47%), Gaps = 0/95 (0%)
Query 45 KEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKL 104
KE + + G + A+E YT+A+ + T RA + K++A A DC+ A+ L
Sbjct 132 KEKGNQFFKDGRFDSAIECYTKAMDADPYNPVPPTNRATCFYRLKKFAVAESDCNLAIAL 191
Query 105 NPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKID 139
+ +AY R L ++A D EM K+D
Sbjct 192 DSKYVKAYIRRAATRTALQKHREALEDYEMVLKLD 226
> hsa:23331 TTC28, KIAA1043; tetratricopeptide repeat domain 28
Length=2481
Score = 46.6 bits (109), Expect = 6e-05, Method: Composition-based stats.
Identities = 28/81 (34%), Positives = 43/81 (53%), Gaps = 0/81 (0%)
Query 48 ATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPD 107
+ +A GD + A+ Y EAL V +LY+ R+ +K ++Y A+ D +A LNP
Sbjct 65 SNQACHDGDFHTAIVLYNEALAVDPQNCILYSNRSAAYMKIQQYDKALDDAIKARLLNPK 124
Query 108 NARAYRIRGTANRYLGHWKQA 128
+AY +G A +YLG A
Sbjct 125 WPKAYFRQGVALQYLGRHADA 145
> mmu:100503562 hypothetical protein LOC100503562
Length=1010
Score = 46.2 bits (108), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 34/112 (30%), Positives = 52/112 (46%), Gaps = 10/112 (8%)
Query 38 YEKLAKAKEAATEAAEAG--------DLNR--AVESYTEALLVGNPTALLYTRRADVLLK 87
+E KA +A E AEA LN+ ++ + AL + Y R
Sbjct 514 FESFTKAVKANPEFAEAFYHRGLCKVKLNKQNSILDFNRALTLNPKHYQAYLSRVAYYGL 573
Query 88 QKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKID 139
+ RY+ AI +C+EA+KL P++ RAY RG Y +K +D+ K+D
Sbjct 574 KGRYSKAILNCNEAIKLYPESVRAYICRGVLKYYNRTYKLGITDLSTAIKMD 625
Score = 32.0 bits (71), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 30/68 (44%), Gaps = 2/68 (2%)
Query 75 ALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEM 134
A LY RA + K+Y A D L L PD A Y +R +G +A SD
Sbjct 940 AALYFNRASFYVYLKKYKLAEEDLGIGLSLKPDEAIMYNLRAQVRGKMGLIAEAMSD--Y 997
Query 135 GQKIDYDE 142
Q +D +E
Sbjct 998 NQALDLEE 1005
Score = 32.0 bits (71), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 20/79 (25%), Positives = 34/79 (43%), Gaps = 0/79 (0%)
Query 55 GDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRI 114
G A+ Y A+ + +L Y ++ L ++++ A ALK NP+N A
Sbjct 852 GQQQSAMADYQAAISLNPSYSLAYFNAGNIYLHHRQFSQASDYFSTALKFNPENEYALMN 911
Query 115 RGTANRYLGHWKQAHSDIE 133
R N L +++A D
Sbjct 912 RAVTNSVLKKYEEAEKDFS 930
Score = 30.0 bits (66), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 28/66 (42%), Gaps = 2/66 (3%)
Query 80 RRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKID 139
+R + + A+ D LKL+P N++A RG A +KQA D I
Sbjct 325 KRGMFYFENGNWIGAVYDFTSLLKLDPYNSKARTYRGRAYFKRNLYKQATQDFSAA--IH 382
Query 140 YDENIW 145
D N W
Sbjct 383 LDPNNW 388
Score = 29.3 bits (64), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 22/90 (24%), Positives = 39/90 (43%), Gaps = 0/90 (0%)
Query 55 GDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRI 114
G ++A+ + EA+ + + Y R + + Y I D A+K++ +N AY
Sbjct 575 GRYSKAILNCNEAIKLYPESVRAYICRGVLKYYNRTYKLGITDLSTAIKMDKNNYTAYYN 634
Query 115 RGTANRYLGHWKQAHSDIEMGQKIDYDENI 144
R +G + A D + +D ENI
Sbjct 635 RALCYTKIGEHQMALRDYGIVLLLDAGENI 664
> ath:AT4G11260 SGT1B; SGT1B; protein binding; K12795 suppressor
of G2 allele of SKP1
Length=358
Score = 45.8 bits (107), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 31/98 (31%), Positives = 52/98 (53%), Gaps = 3/98 (3%)
Query 44 AKEAATEAAEA---GDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDE 100
AKE A +A EA D + AV+ Y++A+ + A + RA +K + A+ D ++
Sbjct 2 AKELAEKAKEAFLDDDFDVAVDLYSKAIDLDPNCAAFFADRAQANIKIDNFTEAVVDANK 61
Query 101 ALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKI 138
A++L P A+AY +GTA L + A + +E G +
Sbjct 62 AIELEPTLAKAYLRKGTACMKLEEYSTAKAALEKGASV 99
> dre:568208 ttc6; tetratricopeptide repeat domain 6
Length=720
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 43/80 (53%), Gaps = 0/80 (0%)
Query 60 AVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTAN 119
V+ + AL + ++ RA + + R+A AI DC+EA+++ P + RA+ +G
Sbjct 259 GVQDFNRALKINPKFYQVHLSRAALYGAEGRHAKAILDCNEAIRIQPKSLRAHLYKGALK 318
Query 120 RYLGHWKQAHSDIEMGQKID 139
YL +K A D+ M +ID
Sbjct 319 FYLKAYKSAVEDLTMAVQID 338
Score = 36.6 bits (83), Expect = 0.064, Method: Compositional matrix adjust.
Identities = 27/98 (27%), Positives = 46/98 (46%), Gaps = 12/98 (12%)
Query 53 EAGDLNRAV------------ESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDE 100
E+ LNRA+ + + EAL + +A Y RA++ +++ +A RD +
Sbjct 620 ESAILNRAITHALLRKVPESLQDFNEALCLNPLSAHAYFNRANLHCSLRQFQSAERDLTQ 679
Query 101 ALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKI 138
AL L P +A Y++R LG ++A D K+
Sbjct 680 ALVLEPGDALLYKLRADVRGCLGWMEEAMEDYRTALKL 717
Score = 29.3 bits (64), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 28/90 (31%), Positives = 43/90 (47%), Gaps = 13/90 (14%)
Query 42 AKAKEAATEAAEAGDLNRAVESYTEALLVGNP-TALLYTRRADVLLKQKRY--AAAIRDC 98
A + + + A D+N A+E NP T L+T R L++Q R ++A++D
Sbjct 526 ATSLQMGNKFAALQDINTAIEY--------NPFTEQLFTNRG--LIQQLRGDKSSAMKDY 575
Query 99 DEALKLNPDNARAYRIRGTANRYLGHWKQA 128
A+ LNP A AY Y G ++QA
Sbjct 576 QTAISLNPAYALAYFNAANLFFYNGQFEQA 605
> ath:AT1G12270 stress-inducible protein, putative
Length=572
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 45/96 (46%), Gaps = 0/96 (0%)
Query 44 AKEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALK 103
AK A +GD A+ +TEA+ + +L++ R+ +YA A+ D E +K
Sbjct 5 AKAKGNAAFSSGDFTTAINHFTEAIALAPTNHVLFSNRSAAHASLHQYAEALSDAKETIK 64
Query 104 LNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKID 139
L P + Y G A+ L ++ A + + G +D
Sbjct 65 LKPYWPKGYSRLGAAHLGLNQFELAVTAYKKGLDVD 100
Score = 32.0 bits (71), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 17/59 (28%), Positives = 31/59 (52%), Gaps = 0/59 (0%)
Query 81 RADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKID 139
+ + K+++Y AI+ EA+K NP++ +AY R + LG + D E ++D
Sbjct 389 KGNDFFKEQKYPEAIKHYTEAIKRNPNDHKAYSNRAASYTKLGAMPEGLKDAEKCIELD 447
> ath:AT4G12400 stress-inducible protein, putative
Length=558
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/96 (28%), Positives = 45/96 (46%), Gaps = 0/96 (0%)
Query 44 AKEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDCDEALK 103
AK A +GD A+ +TEA+ + +LY+ R+ RY A+ D + ++
Sbjct 5 AKSKGNAAFSSGDYATAITHFTEAINLSPTNHILYSNRSASYASLHRYEEALSDAKKTIE 64
Query 104 LNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKID 139
L PD ++ Y G A L + +A + G +ID
Sbjct 65 LKPDWSKGYSRLGAAFIGLSKFDEAVDSYKKGLEID 100
Score = 39.7 bits (91), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 33/64 (51%), Gaps = 0/64 (0%)
Query 39 EKLAKAKEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIRDC 98
EK K K A + D RAVE YT+A+ + + T RA V L+ +Y I DC
Sbjct 228 EKALKEKGEGNVAYKKKDFGRAVEHYTKAMELDDEDISYLTNRAAVYLEMGKYEECIEDC 287
Query 99 DEAL 102
D+A+
Sbjct 288 DKAV 291
Score = 35.8 bits (81), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 29/118 (24%), Positives = 53/118 (44%), Gaps = 9/118 (7%)
Query 81 RADVLLKQKRYAAAIRDCDEALKLNPDNARAYRIRGTANRYLGHWKQAHSDIEMGQKIDY 140
+ + K+++Y A++ EA+K NP++ RAY R LG + D E + I+
Sbjct 375 KGNGFFKEQKYPEAVKHYSEAIKRNPNDVRAYSNRAACYTKLGALPEGLKDAE--KCIEL 432
Query 141 DENIWD-------IQKLVEQKYKIIEEHERAAQRKKEEKERKERERRAREQRANAQRA 191
D + IQ +++ K +E ++ + + +E + RR EQ A R
Sbjct 433 DPSFTKGYSRKGAIQFFMKEYDKAMETYQEGLKHDPKNQEFLDGVRRCVEQINKASRG 490
> xla:432008 wdtc1, MGC78868; WD and tetratricopeptide repeats
1; K11807 WD and tetratricopeptide repeats protein 1
Length=671
Score = 45.4 bits (106), Expect = 1e-04, Method: Composition-based stats.
Identities = 27/75 (36%), Positives = 44/75 (58%), Gaps = 3/75 (4%)
Query 41 LAKAKEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQK---RYAAAIRD 97
L K K+ A +A ++A+E Y+EA+ +A+LY RA +K+K + A+RD
Sbjct 353 LEKVKQKANDAFAQQQWSQAIELYSEAVQRAPRSAMLYGNRAAAYMKRKWDGDHYDALRD 412
Query 98 CDEALKLNPDNARAY 112
C +AL LNP + +A+
Sbjct 413 CLQALALNPAHLKAH 427
> xla:398938 hypothetical protein MGC68614; K11807 WD and tetratricopeptide
repeats protein 1
Length=668
Score = 45.4 bits (106), Expect = 1e-04, Method: Composition-based stats.
Identities = 27/75 (36%), Positives = 44/75 (58%), Gaps = 3/75 (4%)
Query 41 LAKAKEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQK---RYAAAIRD 97
L K K+ A +A ++A+E Y+EA+ +A+LY RA +K+K + A+RD
Sbjct 352 LEKVKQKANDAFAQQQWSQAIELYSEAVQRAPHSAMLYGNRAAAYMKRKWDGDHYDALRD 411
Query 98 CDEALKLNPDNARAY 112
C +AL LNP + +A+
Sbjct 412 CLQALALNPAHLKAH 426
> tgo:TGME49_065380 TPR domain-containing protein ; K09561 STIP1
homology and U-box containing protein 1 [EC:6.3.2.19]
Length=242
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 28/87 (32%), Positives = 40/87 (45%), Gaps = 0/87 (0%)
Query 37 DYEKLAKAKEAATEAAEAGDLNRAVESYTEALLVGNPTALLYTRRADVLLKQKRYAAAIR 96
D + A KE + G AVE YT A+ A+ +T RA K ++ +
Sbjct 2 DATQAAALKERGNLCFKKGMFQSAVELYTRAIECDGSCAVYFTNRALCYKKMGKWTLVLN 61
Query 97 DCDEALKLNPDNARAYRIRGTANRYLG 123
D EA++L DN +AY + G A LG
Sbjct 62 DSREAMQLQKDNVKAYFLMGEALLNLG 88
Lambda K H
0.309 0.126 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5931269072
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40