bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0841_orf1
Length=217
Score E
Sequences producing significant alignments: (Bits) Value
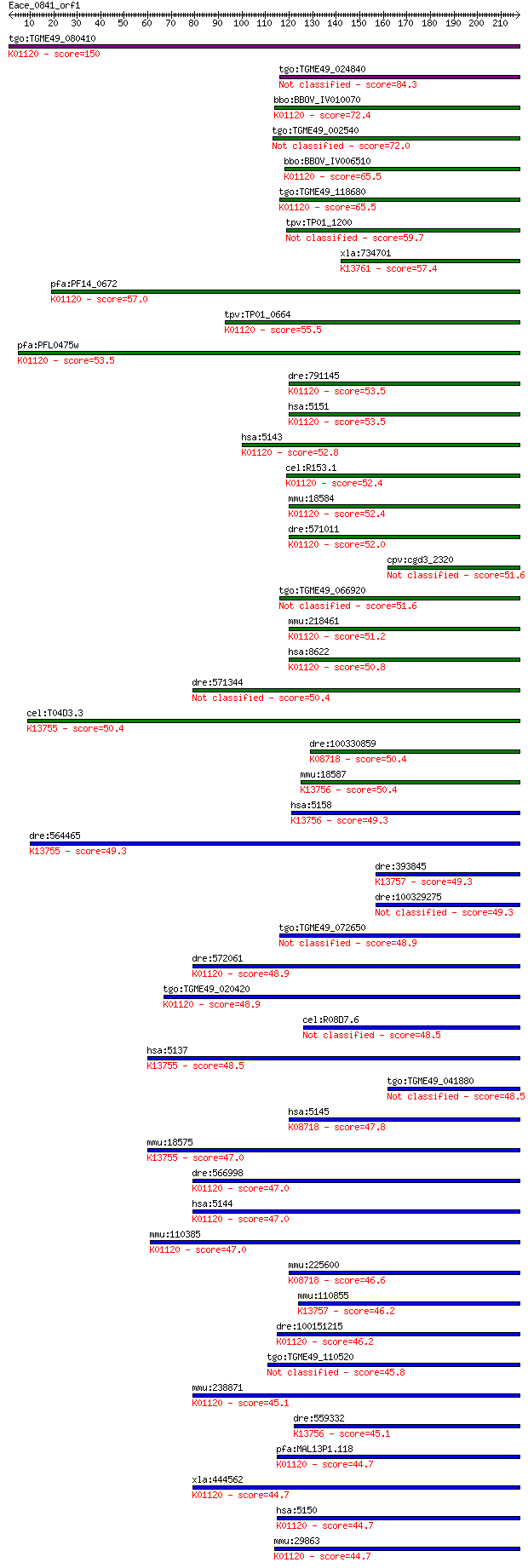
tgo:TGME49_080410 3',5'-cyclic phosphodiesterase, putative (EC... 150 3e-36
tgo:TGME49_024840 cAMP phosphodiesterase, putative (EC:3.1.4.1... 84.3 3e-16
bbo:BBOV_IV010070 23.m06154; 3'5'-cyclic nucleotide phosphodie... 72.4 1e-12
tgo:TGME49_002540 3',5'-cyclic nucleotide phosphodiesterase, p... 72.0 1e-12
bbo:BBOV_IV006510 23.m05980; 3'5'-cyclic nucleotide phosphodie... 65.5 1e-10
tgo:TGME49_118680 3',5'--cyclic-nucleotide phosphodiesterase, ... 65.5 1e-10
tpv:TP01_1200 hypothetical protein 59.7 8e-09
xla:734701 pde9a, MGC116558; phosphodiesterase 9A (EC:3.1.4.17... 57.4 3e-08
pfa:PF14_0672 cyclic nucleotide phosphodiesterase, putative; K... 57.0 5e-08
tpv:TP01_0664 3',5' cyclic nucleotide phosphodiesterase; K0112... 55.5 1e-07
pfa:PFL0475w PDE1; cGMP-specific phosphodiesterase (EC:3.1.4.1... 53.5 5e-07
dre:791145 pde8a, zgc:158458; phosphodiesterase 8A (EC:3.1.4.1... 53.5 5e-07
hsa:5151 PDE8A, FLJ16150, HsT19550; phosphodiesterase 8A (EC:3... 53.5 6e-07
hsa:5143 PDE4C, DPDE1, MGC126222; phosphodiesterase 4C, cAMP-s... 52.8 9e-07
cel:R153.1 pde-4; PhosphoDiEsterase family member (pde-4); K01... 52.4 1e-06
mmu:18584 Pde8a, AI551852, Pde8; phosphodiesterase 8A (EC:3.1.... 52.4 1e-06
dre:571011 IBMX-insensitive phosphodiesterase 8B-like; K01120 ... 52.0 2e-06
cpv:cgd3_2320 cGMP phosphodiesterase A4 51.6 2e-06
tgo:TGME49_066920 phosphodiesterase, putative (EC:3.1.4.17) 51.6 2e-06
mmu:218461 Pde8b, B230331L10Rik, C030047E14Rik; phosphodiester... 51.2 3e-06
hsa:8622 PDE8B, ADSD, FLJ11212, PPNAD3; phosphodiesterase 8B (... 50.8 4e-06
dre:571344 fc68c05, si:ch211-255d18.8, wu:fc68c05; si:ch211-25... 50.4 4e-06
cel:T04D3.3 pde-1; PhosphoDiEsterase family member (pde-1); K1... 50.4 4e-06
dre:100330859 phosphodiesterase 6A-like; K08718 rod cGMP-speci... 50.4 4e-06
mmu:18587 Pde6b, MGC150321, Pdeb, r, rd, rd-1, rd1, rd10; phos... 50.4 5e-06
hsa:5158 PDE6B, CSNB3, CSNBAD2, PDEB, RP40, rd1; phosphodieste... 49.3 1e-05
dre:564465 pde1a; phosphodiesterase 1A, calmodulin-dependent; ... 49.3 1e-05
dre:393845 pde6c, MGC73085, zgc:73085; phosphodiesterase 6C, c... 49.3 1e-05
dre:100329275 phosphodiesterase 6C-like 49.3 1e-05
tgo:TGME49_072650 calcium/calmodulin-dependent 3',5'-cyclic nu... 48.9 1e-05
dre:572061 pde4a; phosphodiesterase 4A, cAMP-specific; K01120 ... 48.9 1e-05
tgo:TGME49_020420 3'5'-cyclic nucleotide phosphodiesterase, pu... 48.9 1e-05
cel:R08D7.6 pde-2; PhosphoDiEsterase family member (pde-2) 48.5 2e-05
hsa:5137 PDE1C, Hcam3; phosphodiesterase 1C, calmodulin-depend... 48.5 2e-05
tgo:TGME49_041880 3', 5'-cyclic nucleotide phosphodiesterase, ... 48.5 2e-05
hsa:5145 PDE6A, CGPR-A, PDEA, RP43; phosphodiesterase 6A, cGMP... 47.8 3e-05
mmu:18575 Pde1c; phosphodiesterase 1C (EC:3.1.4.17); K13755 ca... 47.0 5e-05
dre:566998 novel protein similar to vertebrate phosphodiestera... 47.0 5e-05
hsa:5144 PDE4D, DKFZp686M11213, DPDE3, FLJ97311, HSPDE4D, PDE4... 47.0 6e-05
mmu:110385 Pde4c, Dpde1, E130301F19Rik, MGC31320, dunce; phosp... 47.0 6e-05
mmu:225600 Pde6a, MGC25111, Pdea, nmf282; phosphodiesterase 6A... 46.6 7e-05
mmu:110855 Pde6c, cpfl1; phosphodiesterase 6C, cGMP specific, ... 46.2 8e-05
dre:100151215 phosphodiesterase 7A-like (EC:3.1.4.17); K01120 ... 46.2 9e-05
tgo:TGME49_110520 calcium/calmodulin-dependent 3', 5'-cyclic n... 45.8 1e-04
mmu:238871 Pde4d, 9630011N22Rik, Dpde3; phosphodiesterase 4D, ... 45.1 2e-04
dre:559332 pde6b; phosphodiesterase 6B, cGMP-specific, rod, be... 45.1 2e-04
pfa:MAL13P1.118 3',5'-cyclic nucleotide phosphodiesterase (EC:... 44.7 2e-04
xla:444562 pde4b, MGC83972; phosphodiesterase 4B, cAMP-specifi... 44.7 2e-04
hsa:5150 PDE7A, HCP1, PDE7; phosphodiesterase 7A (EC:3.1.4.17)... 44.7 3e-04
mmu:29863 Pde7b; phosphodiesterase 7B (EC:3.1.4.17); K01120 3'... 44.7 3e-04
> tgo:TGME49_080410 3',5'-cyclic phosphodiesterase, putative (EC:3.1.4.17
1.6.5.3); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=1085
Score = 150 bits (379), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 86/220 (39%), Positives = 128/220 (58%), Gaps = 9/220 (4%)
Query 1 KRIYLPSTPLECVIRNLDLIQRILFNANMEEDEREQVQELLNEARHNLTFTDNIYRFNAD 60
KR TPL+C++ NL L RE++ L+ E + LT ++N+Y+ A+
Sbjct 641 KRRVDTGTPLDCILGNLSEALEALKRPT--PLNREEIFALMQETINMLTTSENVYQVQAE 698
Query 61 TFSDGFARSYVHDLR---REGISRPRKLSRMTSLAARRKSRLFGVVESLSAMCEQSAKIG 117
+ F RS++ DLR RE +SR+ +LA RR+S + G VE+ E +IG
Sbjct 699 YVDNDFTRSFIRDLRMHSREASDF--AVSRLNTLATRRRSVISGRVET--TRTELLEEIG 754
Query 118 TDWSFDCLHFGEISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPYHN 177
W + +PL E+G+ +LSPF+LSP+ L+ +L FL EV Y+ PYHN
Sbjct 755 NSWDVNMFMLRLKLPRPLVEVGYVLLSPFALSPDACLNTQLLRKFLCEVDKAYRNCPYHN 814
Query 178 ALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
+LHG+ VCH+++CLLEML++RE + DLE+ S IA+LCHD
Sbjct 815 SLHGSMVCHLSICLLEMLRLRESLGDLEEASLIIAALCHD 854
> tgo:TGME49_024840 cAMP phosphodiesterase, putative (EC:3.1.4.17
1.1.99.3)
Length=1324
Score = 84.3 bits (207), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 37/102 (36%), Positives = 58/102 (56%), Gaps = 2/102 (1%)
Query 116 IGTDWSFDCLHFGEISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPY 175
+G DW FD LH + + E+G+A+L L P++ L FL + Y++NPY
Sbjct 686 VGVDWDFDMLHLNSQTENVIVEVGYALLC--RLVPDLGCEEVRLIRFLHAIQMQYRDNPY 743
Query 176 HNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
HN +H A V H+T CL ML + M+ ++ ++ +A+LCHD
Sbjct 744 HNKIHSAEVAHLTECLTRMLNAQRNMNSIDKVTLTVAALCHD 785
> bbo:BBOV_IV010070 23.m06154; 3'5'-cyclic nucleotide phosphodiesterase
(EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=485
Score = 72.4 bits (176), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 38/110 (34%), Positives = 62/110 (56%), Gaps = 9/110 (8%)
Query 114 AKIGTDWSFDCL---HFGEISRK-PLCEIGFAILSPFS--LSPEISLHRDVLTAFLEEVT 167
A++G +W+ D +F + R P+ +G ++ P + P+ + ++T L +
Sbjct 146 AQLGKNWNMDFFAMSNFKFVLRSGPVVAVGHQLIDPVGDKIHPDFN---KLITPVLNSIQ 202
Query 168 DCYKENPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
D Y NPYHNALHGA V HM+ LL+ L + +Y++ LE + IA+L HD
Sbjct 203 DVYLPNPYHNALHGACVAHMSCVLLKALSLEQYLTPLEQFAYLIAALGHD 252
> tgo:TGME49_002540 3',5'-cyclic nucleotide phosphodiesterase,
putative (EC:3.1.4.17 1.6.99.5)
Length=1670
Score = 72.0 bits (175), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 36/105 (34%), Positives = 56/105 (53%), Gaps = 2/105 (1%)
Query 113 SAKIGTDWSFDCLHFGEISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKE 172
S + DW+FDCL ++S PL ++G+A+L S ++ L DV+ FL V Y
Sbjct 1277 SEEAARDWNFDCLRHAQLSPTPLVDVGYALL--HRTSEDMRLPPDVVLRFLTAVEIQYNH 1334
Query 173 NPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
PYHN +HG V + L E+L++ + + + +A LCHD
Sbjct 1335 VPYHNCIHGLMVAQKMVALTEVLELSQSIGSRDRALVVVAGLCHD 1379
> bbo:BBOV_IV006510 23.m05980; 3'5'-cyclic nucleotide phosphodiesterase
(EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=777
Score = 65.5 bits (158), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 32/100 (32%), Positives = 53/100 (53%), Gaps = 2/100 (2%)
Query 118 TDWSFDCLHFGEISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPYHN 177
TDW+FD L + + + IG+ +L + + + R V+ FL + + Y+ PYHN
Sbjct 442 TDWNFDVLEYFKKTPAGFMSIGYTLLQKYQ--EDYGIDRSVVVNFLYRIENQYRNVPYHN 499
Query 178 ALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
+HGA V LCL + E++S L++ +A+L HD
Sbjct 500 KMHGAMVAQKVLCLASYTGLLEHLSILDEALLMVAALSHD 539
> tgo:TGME49_118680 3',5'--cyclic-nucleotide phosphodiesterase,
putative (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=812
Score = 65.5 bits (158), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 37/103 (35%), Positives = 54/103 (52%), Gaps = 3/103 (2%)
Query 116 IGTDWSFDCLHFGE-ISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENP 174
+G D S++ L F + L E+G+ +L+ + + VL FL V Y+EN
Sbjct 315 VGIDISYNILEFSKKCPDTVLQEVGYVLLN--RIVCDWGCEDKVLCEFLYMVKTLYRENS 372
Query 175 YHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
YHN +HGA V H +CLL L I M+ L + +A+LCHD
Sbjct 373 YHNQIHGAMVAHYMVCLLRGLGINREMNSLSTAACAVAALCHD 415
> tpv:TP01_1200 hypothetical protein
Length=840
Score = 59.7 bits (143), Expect = 8e-09, Method: Composition-based stats.
Identities = 32/99 (32%), Positives = 46/99 (46%), Gaps = 2/99 (2%)
Query 119 DWSFDCLHFGEISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPYHNA 178
DW+F L + + IG +LS F ++ DV+ +FL V CY YHN
Sbjct 552 DWNFSILDYFRQNPTGFISIGCVLLSEFQ--THFNIPTDVIYSFLGLVEKCYNNVSYHNQ 609
Query 179 LHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
+HG VC LCL + +S ++ I+ LCHD
Sbjct 610 MHGVFVCQKLLCLSNFTNVYSRLSVIDRTILIISGLCHD 648
> xla:734701 pde9a, MGC116558; phosphodiesterase 9A (EC:3.1.4.17);
K13761 high affinity cGMP-specific 3',5'-cyclic phosphodiesterase
9 [EC:3.1.4.35]
Length=342
Score = 57.4 bits (137), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 25/76 (32%), Positives = 45/76 (59%), Gaps = 6/76 (7%)
Query 142 ILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPYHNALHGATVCHMTLCLLEMLQIREYM 201
+++ F + PE + FL V Y+ENP+HN HG +V M C++ Q++E++
Sbjct 63 LVTEFKMEPE------TVRCFLTSVQKHYQENPFHNFYHGFSVTQMMYCVISQCQLQEHL 116
Query 202 SDLEDISACIASLCHD 217
S ++ ++ +A+LCHD
Sbjct 117 SHIDILTLMVAALCHD 132
> pfa:PF14_0672 cyclic nucleotide phosphodiesterase, putative;
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=815
Score = 57.0 bits (136), Expect = 5e-08, Method: Composition-based stats.
Identities = 52/200 (26%), Positives = 94/200 (47%), Gaps = 19/200 (9%)
Query 19 LIQRILFNANMEEDEREQVQELLN-EARHNLTFTDNIYRFNADTFSDGFARSYVHDLRRE 77
+ ++L + N++ED +N + N+ F DNI ++ S+ +L++
Sbjct 319 FLNKLLKSCNIKEDMSSNTSVNINGDTYQNMNFHDNINSNIPKNYN-----SFYEELKK- 372
Query 78 GISRPRKLSRMTSLAARRKSRLFGVVESLSAMCEQSAKIGTDWSFDCLHFGEISRKPLCE 137
L+ L + + ++ ++ C++ K D+SF +G+ S + E
Sbjct 373 ------NLNESDILTIAYEVEVLKNIKKIN--CDEIGK-NWDYSFIDSEYGK-STLVILE 422
Query 138 IGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPYHNALHGATVCHMTLCLLEMLQI 197
+G+ ++SP+ + E L FL + Y NPYHNA HGATVCH++ CL +
Sbjct 423 VGYHLISPYIENNENKKK--KLQLFLLLINSMYFPNPYHNANHGATVCHLSKCLAHITDY 480
Query 198 REYMSDLEDISACIASLCHD 217
Y+++ I IAS+ HD
Sbjct 481 DSYLNNTYMICYLIASIAHD 500
> tpv:TP01_0664 3',5' cyclic nucleotide phosphodiesterase; K01120
3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=450
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 36/131 (27%), Positives = 61/131 (46%), Gaps = 16/131 (12%)
Query 93 ARRKSRLFGVVESLSAMCEQSAKIGTDWSFDCLHFGE----ISRKPLCEIGFAILSPFS- 147
+RRK +L ++ E +IG +W D + + P+ +G +L
Sbjct 91 SRRKKKL-------NSENEYIQQIGLNWHLDLIELSNYPTVLETGPIVVVGKHLLHCVGN 143
Query 148 -LSPEISLHRDVLTAFLEEVTDCYKENPYHNALHGATVCHMTLCLLEMLQIREYMSDLED 206
+ P + + L L+ + Y NPYHNALH ATV HM+ L ++ + ++ E+
Sbjct 144 LIHPNFNTN---LIPVLQNLQQHYLANPYHNALHAATVGHMSKLLSNIVTTKRRLNPYEE 200
Query 207 ISACIASLCHD 217
+ I+SL HD
Sbjct 201 FAFIISSLAHD 211
> pfa:PFL0475w PDE1; cGMP-specific phosphodiesterase (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=954
Score = 53.5 bits (127), Expect = 5e-07, Method: Composition-based stats.
Identities = 53/225 (23%), Positives = 102/225 (45%), Gaps = 18/225 (8%)
Query 5 LPSTPLECVIRNLDLIQRILFNANM-EEDEREQVQELLNEARHNLTFTDNIYRF-NADTF 62
+P++P+E ++ N + IL N+ EE+ + + + + + DNI R N +
Sbjct 503 IPTSPIEDILNNF---KHILETINIIEENPNHNLTTNIMKIKEKIKNCDNILRTKNINQV 559
Query 63 SDGFARSY-----VHDLRREGISRPRKLSRMTSLAARRKSRL----FGVVESL-SAMCEQ 112
G R + + L + ++ P S + +R+ F + SL S+ ++
Sbjct 560 QIGKYRKFEKVYNIWCLDKMYLNYPLNQEETKSFLSNSLNRISFNSFSNMHSLLSSKFQE 619
Query 113 SAKIGTDWSFDCLHFGEISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKE 172
DW+ + + + + IG+ +L P + E + ++ L FL + Y +
Sbjct 620 HYNDIYDWNGNIENIYKAN--TFISIGYKLLYPLGVL-EANFDKEKLKKFLFRICSYYND 676
Query 173 NPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
PYH +LH A V H + +L ML + +S +++ I+SLCHD
Sbjct 677 IPYHTSLHAAQVAHFSKSMLFMLDMNHKISAIDEFCLHISSLCHD 721
> dre:791145 pde8a, zgc:158458; phosphodiesterase 8A (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=817
Score = 53.5 bits (127), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 51/100 (51%), Gaps = 2/100 (2%)
Query 120 WSFDCLHFGEIS-RKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCY-KENPYHN 177
W FD + + ++PL +G + S F + + L ++L+ + Y NPYHN
Sbjct 484 WDFDIFNLESATLKRPLAFLGLKLFSRFGVCEFLGCAEATLRSWLQNIEASYHGSNPYHN 543
Query 178 ALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
+ H A V H T L ++++ + +++++A IA+ HD
Sbjct 544 STHAADVLHATAYFLCKERVKQSLDPIDEVAALIAAAVHD 583
> hsa:5151 PDE8A, FLJ16150, HsT19550; phosphodiesterase 8A (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=829
Score = 53.5 bits (127), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 48/100 (48%), Gaps = 2/100 (2%)
Query 120 WSFDCLHF-GEISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCY-KENPYHN 177
W FD +PL +G + + F + + L ++L+ + Y NPYHN
Sbjct 498 WDFDIFELEAATHNRPLIYLGLKMFARFGICEFLHCSESTLRSWLQIIEANYHSSNPYHN 557
Query 178 ALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
+ H A V H T L +I+E + +++++A IA+ HD
Sbjct 558 STHSADVLHATAYFLSKERIKETLDPIDEVAALIAATIHD 597
> hsa:5143 PDE4C, DPDE1, MGC126222; phosphodiesterase 4C, cAMP-specific
(EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=712
Score = 52.8 bits (125), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 36/120 (30%), Positives = 49/120 (40%), Gaps = 2/120 (1%)
Query 100 FGVVESLSAMCEQSAKIGTDWSFDCLHFGEIS-RKPLCEIGFAILSPFSLSPEISLHRDV 158
FGV + + W D E+S +PL I F+I L + D
Sbjct 310 FGVQTDQEEQLAKELEDTNKWGLDVFKVAELSGNRPLTAIIFSIFQERDLLKTFQIPADT 369
Query 159 LTAFLEEVTDCYKEN-PYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
L +L + Y N YHN+LH A V T LL + +DLE ++A AS HD
Sbjct 370 LATYLLMLEGHYHANVAYHNSLHAADVAQSTHVLLATPALEAVFTDLEILAALFASAIHD 429
> cel:R153.1 pde-4; PhosphoDiEsterase family member (pde-4); K01120
3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=673
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/100 (30%), Positives = 46/100 (46%), Gaps = 1/100 (1%)
Query 119 DWSFDCLHFGEISRK-PLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPYHN 177
DW D E+S+ L + F++L +L +H+ L +L + Y+ N YHN
Sbjct 348 DWGPDVFKIDELSKNHSLTVVTFSLLRQRNLFKTFEIHQSTLVTYLLNLEHHYRNNHYHN 407
Query 178 ALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
+H A V LL + E +DLE ++A A HD
Sbjct 408 FIHAADVAQSMHVLLMSPVLTEVFTDLEVLAAIFAGAVHD 447
> mmu:18584 Pde8a, AI551852, Pde8; phosphodiesterase 8A (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=823
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 48/100 (48%), Gaps = 2/100 (2%)
Query 120 WSFDCLHFGEISR-KPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCY-KENPYHN 177
W FD ++ +PL +G + F + + L ++ + + Y NPYHN
Sbjct 493 WDFDIFELEVATQNRPLIYLGLKTFARFGMCEFLQCSETTLRSWFQMIESNYHSSNPYHN 552
Query 178 ALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
+ H A V H T L +I+E + +++++A IA+ HD
Sbjct 553 STHAADVLHATAYFLSRDKIKETLDRIDEVAALIAATVHD 592
> dre:571011 IBMX-insensitive phosphodiesterase 8B-like; K01120
3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=784
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 50/100 (50%), Gaps = 2/100 (2%)
Query 120 WSFDCLHF-GEISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEV-TDCYKENPYHN 177
W F+ L ++PL +G I S F + ++ L +L+ + T+ + N YHN
Sbjct 450 WEFNILELEAATHKRPLSYLGLKIFSAFGVCEFLNCSESTLRLWLQVIETNYHSSNSYHN 509
Query 178 ALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
+ H A V H T L +++ + L++++A +A+ HD
Sbjct 510 STHAADVLHATAYFLRKERVKASLDQLDEVAALLAATVHD 549
> cpv:cgd3_2320 cGMP phosphodiesterase A4
Length=997
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 22/56 (39%), Positives = 32/56 (57%), Gaps = 0/56 (0%)
Query 162 FLEEVTDCYKENPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
F E+ Y N YHN LHG VCH+ +CL + ++ +E +++ IASL HD
Sbjct 711 FFRELNIRYLNNKYHNELHGTNVCHLAICLSRATGLWSHLDTVERLASVIASLGHD 766
> tgo:TGME49_066920 phosphodiesterase, putative (EC:3.1.4.17)
Length=1065
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 31/102 (30%), Positives = 45/102 (44%), Gaps = 2/102 (1%)
Query 116 IGTDWSFDCLHFGEISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPY 175
+G W+FD L E + L GF +L+ + + LT FL ++ NPY
Sbjct 556 VGVAWNFDLLEVEEETGHALSLTGFVLLN--GVVADWGCPPSRLTNFLLSCEAQHQANPY 613
Query 176 HNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
HN H A V H T L + +E + +A+LCHD
Sbjct 614 HNQRHAAMVAHATAWLANTVGALNKCDSVERATLYVAALCHD 655
> mmu:218461 Pde8b, B230331L10Rik, C030047E14Rik; phosphodiesterase
8B (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=788
Score = 51.2 bits (121), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 49/100 (49%), Gaps = 2/100 (2%)
Query 120 WSFDCLHFGEISRK-PLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCY-KENPYHN 177
W F+ ++ K PL +G + S F + ++ L A+L+ + Y N YHN
Sbjct 460 WDFNIFELEAVTHKRPLVYLGLKVFSRFGVCEFLNCTETTLRAWLQVIEANYHSSNAYHN 519
Query 178 ALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
+ H A V H T L +++ + L++++A IA+ HD
Sbjct 520 STHAADVLHATAFFLGKERVKGSLDQLDEVAALIAATVHD 559
> hsa:8622 PDE8B, ADSD, FLJ11212, PPNAD3; phosphodiesterase 8B
(EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=788
Score = 50.8 bits (120), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 48/100 (48%), Gaps = 2/100 (2%)
Query 120 WSFDCLHFGEISRK-PLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCY-KENPYHN 177
W F+ I+ K PL +G + S F + ++ L A+ + + Y N YHN
Sbjct 460 WDFNIFELEAITHKRPLVYLGLKVFSRFGVCEFLNCSETTLRAWFQVIEANYHSSNAYHN 519
Query 178 ALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
+ H A V H T L +++ + L++++A IA+ HD
Sbjct 520 STHAADVLHATAFFLGKERVKGSLDQLDEVAALIAATVHD 559
> dre:571344 fc68c05, si:ch211-255d18.8, wu:fc68c05; si:ch211-255d18.1
Length=709
Score = 50.4 bits (119), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 42/141 (29%), Positives = 58/141 (41%), Gaps = 3/141 (2%)
Query 79 ISRPRKLSRMTSLAARRKSRLFGVVESLSAMCEQSAKIGTDWSFDCLHFGEIS-RKPLCE 137
IS RKLS SL R FGV ++ + + W D E S +PL
Sbjct 262 ISGVRKLSPSPSLPPTCIPR-FGVNTQHESLLAKELEDLDRWGIDIFKIAEYSGNRPLTV 320
Query 138 IGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKEN-PYHNALHGATVCHMTLCLLEMLQ 196
+ + I L + D FL + D Y + YHN +H A V T LL
Sbjct 321 MMYTIFQERDLLKTFKIPSDTFLTFLMTLEDHYHADVAYHNNIHAADVVQSTHVLLSTPA 380
Query 197 IREYMSDLEDISACIASLCHD 217
+ + +DLE ++A AS HD
Sbjct 381 LEDVFTDLEIMAALFASAIHD 401
> cel:T04D3.3 pde-1; PhosphoDiEsterase family member (pde-1);
K13755 calcium/calmodulin-dependent 3',5'-cyclic nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=664
Score = 50.4 bits (119), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 52/226 (23%), Positives = 90/226 (39%), Gaps = 34/226 (15%)
Query 9 PLECVIRNLDLIQRILFNANMEE-----DEREQVQELLNEARHNLTFTDNIYRFNADTFS 63
PLE + RN++ +L A M+E DE + + E+ E T D + + A TF+
Sbjct 166 PLEDLKRNIEYAALVLETAYMDETRRICDEDDDLAEVTPE-----TVPDEVREWLAATFT 220
Query 64 DGFARSYVHDLRREGISRPRKLSRMTSLAARRKSRLF--------GVVE-SLSAMCEQSA 114
R+ + R + S+A ++ +F VV+ + +
Sbjct 221 ------------RQNAGKKRDKPKFKSVANAIRTGIFFEKLFRKQQVVQCPIPPEIAELM 268
Query 115 KIGTDWSFDCLHFGEISRK-PLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKE- 172
K WSF E+S L +GF + + + + L +L + Y +
Sbjct 269 KEVCTWSFSPFQLNEVSEGHALKYVGFELFNRYGFMDRFKVPLTALENYLSALEVGYSKH 328
Query 173 -NPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
NPYHN +H A V + +L + + DLE ++ +L HD
Sbjct 329 NNPYHNVVHAADVTQSSHFMLSQTGLANSLGDLELLAVLFGALIHD 374
> dre:100330859 phosphodiesterase 6A-like; K08718 rod cGMP-specific
3',5'-cyclic phosphodiesterase subunit alpha [EC:3.1.4.35]
Length=858
Score = 50.4 bits (119), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 41/89 (46%), Gaps = 0/89 (0%)
Query 129 EISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPYHNALHGATVCHMT 188
E S L + G + + + + R+ L F+ V+ Y++ YHN HG V
Sbjct 513 EFSELDLVKCGIKMYYELGVVDKFHIPRETLVRFVYSVSKGYRKITYHNWRHGFNVGQTM 572
Query 189 LCLLEMLQIREYMSDLEDISACIASLCHD 217
LL ++ Y +DLE ++ A LCHD
Sbjct 573 FTLLMTGDLKRYYTDLEAMAMVTAGLCHD 601
> mmu:18587 Pde6b, MGC150321, Pdeb, r, rd, rd-1, rd1, rd10; phosphodiesterase
6B, cGMP, rod receptor, beta polypeptide (EC:3.1.4.35);
K13756 rod cGMP-specific 3',5'-cyclic phosphodiesterase
subunit beta [EC:3.1.4.35]
Length=856
Score = 50.4 bits (119), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 27/95 (28%), Positives = 44/95 (46%), Gaps = 2/95 (2%)
Query 125 LHFGEI--SRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPYHNALHGA 182
HF ++ + L + G + + + + ++VL FL V+ Y+ YHN HG
Sbjct 504 FHFSDLECTELELVKCGIQMYYELGVVRKFQIPQEVLVRFLFSVSKAYRRITYHNWRHGF 563
Query 183 TVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
V LL +++ Y +DLE + A LCHD
Sbjct 564 NVAQTMFTLLMTGKLKSYYTDLEAFAMVTAGLCHD 598
> hsa:5158 PDE6B, CSNB3, CSNBAD2, PDEB, RP40, rd1; phosphodiesterase
6B, cGMP-specific, rod, beta (EC:3.1.4.35); K13756 rod
cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta
[EC:3.1.4.35]
Length=854
Score = 49.3 bits (116), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 28/101 (27%), Positives = 47/101 (46%), Gaps = 4/101 (3%)
Query 121 SFDC--LHFGEI--SRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPYH 176
+FD HF ++ + L + G + + + + ++VL FL ++ Y+ YH
Sbjct 498 TFDIYEFHFSDLECTELDLVKCGIQMYYELGVVRKFQIPQEVLVRFLFSISKGYRRITYH 557
Query 177 NALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
N HG V LL +++ Y +DLE + A LCHD
Sbjct 558 NWRHGFNVAQTMFTLLMTGKLKSYYTDLEAFAMVTAGLCHD 598
> dre:564465 pde1a; phosphodiesterase 1A, calmodulin-dependent;
K13755 calcium/calmodulin-dependent 3',5'-cyclic nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=625
Score = 49.3 bits (116), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 53/228 (23%), Positives = 94/228 (41%), Gaps = 22/228 (9%)
Query 10 LECVIRNLDL--IQRILFNANMEEDEREQVQELLNEARHNLTFTDNIYRFNADTFS---- 63
L C+++ LD + + N+E ++E R L D + AD+
Sbjct 38 LRCLVKQLDKGEVNVVDLRKNIEYAASVLEAVYIDETRRLLDTEDELSDIQADSVPMEVR 97
Query 64 DGFARSYVHDL---RREGISRPRKLSRMTSLAA--------RRKSRLFGVVESLSAMCEQ 112
D A ++ + RR +PR S + ++ A RR S + G+ S +
Sbjct 98 DWLASTFTRKMGVVRRRPEEKPRFRSIVHAVQAGIFVERMYRRTSNMAGLTYPPSVIT-- 155
Query 113 SAKIGTDWSFDCLHFGEISRK-PLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYK 171
+ K WSFD E S L + + +L+ + L + L +F+E + Y
Sbjct 156 ALKDVDKWSFDVFKLQEASSDHALKFLVYELLTRYDLISRFRIPVSSLVSFVEALEIGYS 215
Query 172 E--NPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
+ NPYHN +H A V L+ + ++++LE ++ A+ HD
Sbjct 216 KHRNPYHNLIHAADVTQTAHYLMLHTGVMHWLTELEILAMVFAAAIHD 263
> dre:393845 pde6c, MGC73085, zgc:73085; phosphodiesterase 6C,
cGMP-specific, cone, alpha prime (EC:3.1.4.17); K13757 cone
cGMP-specific 3',5'-cyclic phosphodiesterase subunit alpha'
[EC:3.1.4.35]
Length=852
Score = 49.3 bits (116), Expect = 1e-05, Method: Composition-based stats.
Identities = 22/61 (36%), Positives = 34/61 (55%), Gaps = 0/61 (0%)
Query 157 DVLTAFLEEVTDCYKENPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCH 216
+VLT ++ V Y++ YHN HG V CLL+ ++R Y +DL+ + A+ CH
Sbjct 538 EVLTRWMYTVRKGYRDITYHNWRHGFNVGQTMFCLLQTGKLRRYYTDLDAFAMVAAAFCH 597
Query 217 D 217
D
Sbjct 598 D 598
> dre:100329275 phosphodiesterase 6C-like
Length=852
Score = 49.3 bits (116), Expect = 1e-05, Method: Composition-based stats.
Identities = 22/61 (36%), Positives = 34/61 (55%), Gaps = 0/61 (0%)
Query 157 DVLTAFLEEVTDCYKENPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCH 216
+VLT ++ V Y++ YHN HG V CLL+ ++R Y +DL+ + A+ CH
Sbjct 538 EVLTRWMYTVRKGYRDITYHNWRHGFNVGQTMFCLLQTGKLRRYYTDLDAFAMVAAAFCH 597
Query 217 D 217
D
Sbjct 598 D 598
> tgo:TGME49_072650 calcium/calmodulin-dependent 3',5'-cyclic
nucleotide phosphodiesterase, putative (EC:3.1.4.17)
Length=1320
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 30/115 (26%), Positives = 51/115 (44%), Gaps = 13/115 (11%)
Query 116 IGTDWSFDCLHF-GEISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENP 174
+G WS D E + L +G+ +L+P + ++ R+V+ FL + Y + P
Sbjct 990 VGILWSLDLFKLDKECNGNALLHVGYQLLAPLLAAGYLTCSREVVLDFLYSLQCLYIDTP 1049
Query 175 YHNALHGATVCHMTLCLLEMLQIREYMSDLED------------ISACIASLCHD 217
YHN LH A+V H+ + L + + + +S IA+L HD
Sbjct 1050 YHNQLHAASVAHLAFSICHFLDLFAPLENAPGDAAASQPVFAQYLSLAIAALGHD 1104
> dre:572061 pde4a; phosphodiesterase 4A, cAMP-specific; K01120
3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=736
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 45/142 (31%), Positives = 63/142 (44%), Gaps = 5/142 (3%)
Query 79 ISRPRKLSRMTSLAARRKSRLFGV-VESLSAMCEQSAKIGTDWSFDCLHFGEISR-KPLC 136
IS +KL+ +SL + R FGV E A+ + + W + H E S +PL
Sbjct 294 ISGVKKLTHSSSLTSAALPR-FGVKTEQEDALARELNDLNK-WGLNIFHVAEFSNNRPLS 351
Query 137 EIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKEN-PYHNALHGATVCHMTLCLLEML 195
I FAI L + D ++ + D Y N YHN+LH A V T LL
Sbjct 352 CIMFAIFQERDLLKTFRIPVDTFITYVMTLEDHYHANVAYHNSLHAADVTQSTHVLLSTP 411
Query 196 QIREYMSDLEDISACIASLCHD 217
+ SDLE ++A A+ HD
Sbjct 412 ALDAVFSDLEILAALFAAAIHD 433
> tgo:TGME49_020420 3'5'-cyclic nucleotide phosphodiesterase,
putative (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=466
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 40/152 (26%), Positives = 56/152 (36%), Gaps = 38/152 (25%)
Query 67 ARSYVHDLRREGISRPRKLSRMTSLAARRKSRLFGVVESLSAMCEQSAKIGTDWSFDCLH 126
+R++ D E R+ S +T A LFG+VE ++G DW D
Sbjct 85 SRTWAKDPPSEAKPLARRHSFVTLGAI--DDNLFGLVE----------RVGEDWDLDMFE 132
Query 127 F-GEISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPYHNALHGATVC 185
+ +R L GF + Y+ NPYHN HGA V
Sbjct 133 LCKQTNRMTLMATGFRLQQ-------------------------YRHNPYHNEQHGAAVA 167
Query 186 HMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
HM + LL Q L + +A+L HD
Sbjct 168 HMMVFLLRACQAWHMFKPLYQTAIIVAALVHD 199
> cel:R08D7.6 pde-2; PhosphoDiEsterase family member (pde-2)
Length=856
Score = 48.5 bits (114), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/92 (32%), Positives = 43/92 (46%), Gaps = 8/92 (8%)
Query 126 HFGEISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPYHNALHGATVC 185
HF S ++GF++L L++ L+ + V+ Y+ PYHN H V
Sbjct 377 HFHRASMMFFEDLGFSML--------YKLNKRKLSYLVLRVSAGYRPVPYHNWSHAFAVT 428
Query 186 HMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
H L IR +SD+E +S IA LCHD
Sbjct 429 HFCWLTLRTDAIRRALSDMERLSLLIACLCHD 460
> hsa:5137 PDE1C, Hcam3; phosphodiesterase 1C, calmodulin-dependent
70kDa (EC:3.1.4.17); K13755 calcium/calmodulin-dependent
3',5'-cyclic nucleotide phosphodiesterase [EC:3.1.4.17]
Length=634
Score = 48.5 bits (114), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 45/169 (26%), Positives = 71/169 (42%), Gaps = 14/169 (8%)
Query 60 DTFSDGFARSYVHDLRREGISRPRKLSRMTSLAA--------RRKSRLFGVVESLSAMCE 111
D + F R LRR +PR S + ++ A RR S + G+ S
Sbjct 104 DWLASTFTRQMGMMLRRSD-EKPRFKSIVHAVQAGIFVERMYRRTSNMVGL--SYPPAVI 160
Query 112 QSAKIGTDWSFDCLHFGEISRK-PLCEIGFAILSPFSLSPEISLHRDVLTAFLE--EVTD 168
++ K WSFD E S L I + +L+ + L + L +F+E EV
Sbjct 161 EALKDVDKWSFDVFSLNEASGDHALKFIFYELLTRYDLISRFKIPISALVSFVEALEVGY 220
Query 169 CYKENPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
+NPYHN +H A V LL + ++++LE + ++ HD
Sbjct 221 SKHKNPYHNLMHAADVTQTVHYLLYKTGVANWLTELEIFAIIFSAAIHD 269
> tgo:TGME49_041880 3', 5'-cyclic nucleotide phosphodiesterase,
putative (EC:2.1.1.43 3.1.4.17)
Length=1281
Score = 48.5 bits (114), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 33/56 (58%), Gaps = 0/56 (0%)
Query 162 FLEEVTDCYKENPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
F++ CY+ YHN LHGA V H T+ L L++ ++ E ++ +A+LCHD
Sbjct 850 FMKVAQMCYQPTIYHNHLHGAQVAHNTVWLARKLELASSLTAPELVALIVAALCHD 905
> hsa:5145 PDE6A, CGPR-A, PDEA, RP43; phosphodiesterase 6A, cGMP-specific,
rod, alpha (EC:3.1.4.35); K08718 rod cGMP-specific
3',5'-cyclic phosphodiesterase subunit alpha [EC:3.1.4.35]
Length=860
Score = 47.8 bits (112), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 25/103 (24%), Positives = 48/103 (46%), Gaps = 8/103 (7%)
Query 120 WSFDCLHFGEISRKPLCEI-----GFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENP 174
+ + HF ++ PL E+ G + + + + ++ L F+ ++ Y++
Sbjct 501 YEINKFHFSDL---PLTELELVKCGIQMYYELKVVDKFHIPQEALVRFMYSLSKGYRKIT 557
Query 175 YHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
YHN HG V LL +++ Y +DLE ++ A+ CHD
Sbjct 558 YHNWRHGFNVGQTMFSLLVTGKLKRYFTDLEALAMVTAAFCHD 600
> mmu:18575 Pde1c; phosphodiesterase 1C (EC:3.1.4.17); K13755
calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=631
Score = 47.0 bits (110), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 45/169 (26%), Positives = 70/169 (41%), Gaps = 14/169 (8%)
Query 60 DTFSDGFARSYVHDLRREGISRPRKLSRMTSLAA--------RRKSRLFGVVESLSAMCE 111
D + F R LRR +PR S + ++ A RR S + G+ S
Sbjct 104 DWLASTFTRQMGMMLRRSD-EKPRFKSIVHAVQAGIFVERMYRRTSNMVGL--SYPPAVI 160
Query 112 QSAKIGTDWSFDCLHFGEISRK-PLCEIGFAILSPFSLSPEISLHRDVLTAFLE--EVTD 168
+ K WSFD E S L I + +L+ + L + L +F+E EV
Sbjct 161 DALKDVDTWSFDVFSLNEASGDHALKFIFYELLTRYDLISRFKIPISALVSFVEALEVGY 220
Query 169 CYKENPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
+NPYHN +H A V LL + ++++LE + ++ HD
Sbjct 221 SKHKNPYHNLMHAADVTQTVHYLLYKTGVANWLTELEIFAIIFSAAIHD 269
> dre:566998 novel protein similar to vertebrate phosphodiesterase
4D, cAMP-specific (phosphodiesterase E3 dunce homolog,
Drosophila) (PDE4D); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=745
Score = 47.0 bits (110), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 38/141 (26%), Positives = 56/141 (39%), Gaps = 3/141 (2%)
Query 79 ISRPRKLSRMTSLAARRKSRLFGVVESLSAMCEQSAKIGTDWSFDCLHFGEIS-RKPLCE 137
IS +KL+ +SL R FGV + + W + E S +PL
Sbjct 276 ISGVKKLTHSSSLTNSNIPR-FGVKTETEDELAKELEDVNKWGLNVFKVSEFSGNRPLTV 334
Query 138 IGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKEN-PYHNALHGATVCHMTLCLLEMLQ 196
+ + I L + D +L + D Y + YHN +H A V T LL
Sbjct 335 MMYTIFQERDLLKTFKIPLDTFITYLMTLEDHYHADVAYHNNIHAADVTQSTHVLLSTPA 394
Query 197 IREYMSDLEDISACIASLCHD 217
+ +DLE ++A AS HD
Sbjct 395 LEAVFTDLEILAAIFASAIHD 415
> hsa:5144 PDE4D, DKFZp686M11213, DPDE3, FLJ97311, HSPDE4D, PDE4DN2,
STRK1; phosphodiesterase 4D, cAMP-specific (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=809
Score = 47.0 bits (110), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 40/141 (28%), Positives = 56/141 (39%), Gaps = 3/141 (2%)
Query 79 ISRPRKLSRMTSLAARRKSRLFGVVESLSAMCEQSAKIGTDWSFDCLHFGEIS-RKPLCE 137
IS +KL +SL R FGV + + + W E+S +PL
Sbjct 364 ISGVKKLMHSSSLTNSSIPR-FGVKTEQEDVLAKELEDVNKWGLHVFRIAELSGNRPLTV 422
Query 138 IGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKEN-PYHNALHGATVCHMTLCLLEMLQ 196
I I L + D L +L + D Y + YHN +H A V T LL
Sbjct 423 IMHTIFQERDLLKTFKIPVDTLITYLMTLEDHYHADVAYHNNIHAADVVQSTHVLLSTPA 482
Query 197 IREYMSDLEDISACIASLCHD 217
+ +DLE ++A AS HD
Sbjct 483 LEAVFTDLEILAAIFASAIHD 503
> mmu:110385 Pde4c, Dpde1, E130301F19Rik, MGC31320, dunce; phosphodiesterase
4C, cAMP specific (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=652
Score = 47.0 bits (110), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 41/159 (25%), Positives = 62/159 (38%), Gaps = 12/159 (7%)
Query 61 TFSDGFARSYVHDLRREGISRPRKLSRMTSLAARRKSRLFGVVESLSAMCEQSAKIGTDW 120
T D + + + +LRR S TSL R FGV + + W
Sbjct 282 TEDDPWPMAQITELRR---------SSHTSLPTAAIPR-FGVQTDQEEQLAKELEDTNKW 331
Query 121 SFDCLHFGEIS-RKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKEN-PYHNA 178
D E+S +PL + F++ L + D L A+L + Y + YHN+
Sbjct 332 GLDVFKVAELSGNRPLTAVIFSVFQERDLLKTFQIPADTLLAYLLTLEGHYHSDVAYHNS 391
Query 179 LHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
+H A V LL + +DLE ++A A HD
Sbjct 392 MHAADVVQSAHVLLGTPALEAVFTDLEVLAAIFACAIHD 430
> mmu:225600 Pde6a, MGC25111, Pdea, nmf282; phosphodiesterase
6A, cGMP-specific, rod, alpha (EC:3.1.4.35); K08718 rod cGMP-specific
3',5'-cyclic phosphodiesterase subunit alpha [EC:3.1.4.35]
Length=860
Score = 46.6 bits (109), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 25/103 (24%), Positives = 47/103 (45%), Gaps = 8/103 (7%)
Query 120 WSFDCLHFGEISRKPLCEI-----GFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENP 174
+ + HF ++ PL E+ G + + + + ++ L F+ ++ Y+
Sbjct 501 YEINKFHFSDL---PLTELELVKCGIQMYYELRVVDKFHIPQEALVRFMYSLSKGYRRIT 557
Query 175 YHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
YHN HG V LL +++ Y +DLE ++ A+ CHD
Sbjct 558 YHNWRHGFNVGQTMFSLLVTGKLKRYFTDLEALAMVTAAFCHD 600
> mmu:110855 Pde6c, cpfl1; phosphodiesterase 6C, cGMP specific,
cone, alpha prime (EC:3.1.4.35); K13757 cone cGMP-specific
3',5'-cyclic phosphodiesterase subunit alpha' [EC:3.1.4.35]
Length=836
Score = 46.2 bits (108), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 25/94 (26%), Positives = 43/94 (45%), Gaps = 0/94 (0%)
Query 124 CLHFGEISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPYHNALHGAT 183
C I+ L + G + ++ + + +VLT ++ V Y+ YHN HG
Sbjct 510 CFSDFPITEHELVKCGLRLFLEINVVEKFKVPVEVLTRWMYTVRKGYRPVTYHNWRHGFN 569
Query 184 VCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
V LL ++++Y +DLE + A+ CHD
Sbjct 570 VGQTMFTLLMTGRLKKYYTDLEAFAMLAAAFCHD 603
> dre:100151215 phosphodiesterase 7A-like (EC:3.1.4.17); K01120
3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=490
Score = 46.2 bits (108), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 30/105 (28%), Positives = 51/105 (48%), Gaps = 3/105 (2%)
Query 115 KIGTDWSFDCLHFGEISR-KPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVT-DCYKE 172
K+G+ W+FD F ++ L + F +LS + L L + FL V D + +
Sbjct 152 KVGS-WNFDIFLFDRLTNGNSLVFLTFHLLSQYGLIELFQLDMVKVRRFLVLVQEDYHNQ 210
Query 173 NPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
NPYHNA+H A V C L ++ + ++ + + +A+ HD
Sbjct 211 NPYHNAVHAADVTQAMHCYLREPKLAQSLTSFDILLGLLAAATHD 255
> tgo:TGME49_110520 calcium/calmodulin-dependent 3', 5'-cyclic
nucleotide phosphodiesterase, putative (EC:3.4.21.69 3.1.4.17)
Length=1531
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 36/112 (32%), Positives = 52/112 (46%), Gaps = 13/112 (11%)
Query 111 EQSAKIGTDWSFDCLHFGEISRKPLCEIGFAILSPFSLSPEI-SLH----RDVLTAFLEE 165
E IG W FD L F +S PL + L PEI SLH ++V+ L+
Sbjct 1155 EAQMAIGQVWDFDMLSFAALSPNPL-----VEVGLVLLLPEIDSLHCCTDQEVIV-LLQN 1208
Query 166 VTDCYKENPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
+ Y +NPYH+ +H A V H CL+ L + S ++ +A+ HD
Sbjct 1209 LQARYLQNPYHSQVHAAEVVHTAACLMRCLIPQR--SAFANLCTLVAAAAHD 1258
> mmu:238871 Pde4d, 9630011N22Rik, Dpde3; phosphodiesterase 4D,
cAMP specific (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=747
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 40/141 (28%), Positives = 56/141 (39%), Gaps = 3/141 (2%)
Query 79 ISRPRKLSRMTSLAARRKSRLFGVVESLSAMCEQSAKIGTDWSFDCLHFGEIS-RKPLCE 137
IS +KL +SL R FGV + + + W E+S +PL
Sbjct 303 ISGVKKLMHSSSLTNSCIPR-FGVKTEQEDVLAKELEDVNKWGLHVFRIAELSGNRPLTV 361
Query 138 IGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKEN-PYHNALHGATVCHMTLCLLEMLQ 196
I I L + D L +L + D Y + YHN +H A V T LL
Sbjct 362 IMHTIFQERDLLKTFKIPVDTLITYLMTLEDHYHADVAYHNNIHAADVVQSTHVLLSTPA 421
Query 197 IREYMSDLEDISACIASLCHD 217
+ +DLE ++A AS HD
Sbjct 422 LEAVFTDLEILAAIFASAIHD 442
> dre:559332 pde6b; phosphodiesterase 6B, cGMP-specific, rod,
beta; K13756 rod cGMP-specific 3',5'-cyclic phosphodiesterase
subunit beta [EC:3.1.4.35]
Length=854
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/96 (26%), Positives = 41/96 (42%), Gaps = 6/96 (6%)
Query 122 FDCLHFGEISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPYHNALHG 181
FDC L G + + + + ++VL F+ V+ Y+ YHN HG
Sbjct 510 FDCTEL------QLVMCGIQMYYEVGVVKKFQVPQEVLVRFMYSVSKGYRRITYHNWRHG 563
Query 182 ATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
V LL ++++Y +DLE ++ A HD
Sbjct 564 FNVGQTMFTLLTTGKLKQYYTDLEVMAMITAGFLHD 599
> pfa:MAL13P1.118 3',5'-cyclic nucleotide phosphodiesterase (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=1139
Score = 44.7 bits (104), Expect = 2e-04, Method: Composition-based stats.
Identities = 27/103 (26%), Positives = 49/103 (47%), Gaps = 4/103 (3%)
Query 115 KIGTDWSFDCLHFGEISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENP 174
K+ +DW F+CL + + S P +I +++ + ++ +++ FL V Y P
Sbjct 790 KLLSDWDFNCLTYFDESEYPFFDINLSLICTI----DHNIPINIIINFLCFVEKQYNNVP 845
Query 175 YHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
YHN +H V CL + L I + + + I+ +CHD
Sbjct 846 YHNTIHATMVTQKFFCLAKKLGIYDDLEYKIKLVMFISGICHD 888
> xla:444562 pde4b, MGC83972; phosphodiesterase 4B, cAMP-specific;
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=721
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 41/141 (29%), Positives = 58/141 (41%), Gaps = 3/141 (2%)
Query 79 ISRPRKLSRMTSLAARRKSRLFGVVESLSAMCEQSAKIGTDWSFDCLHFGEIS-RKPLCE 137
IS +KL +SL SR FGV M + + W + S +PL
Sbjct 294 ISGVKKLKHSSSLNNTSMSR-FGVKTDYEDMLSKELEDLNKWGLNIFKVASYSCNRPLTC 352
Query 138 IGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKEN-PYHNALHGATVCHMTLCLLEMLQ 196
I +AI L + D L + + D Y + YHN+LH A V T LL
Sbjct 353 IMYAIFQERDLLKTFKIPVDTLITYTMTLEDHYHSDVAYHNSLHAADVTQSTHVLLSTPA 412
Query 197 IREYMSDLEDISACIASLCHD 217
+ +DLE ++A A+ HD
Sbjct 413 LDAVFTDLEILAAIFAAAIHD 433
> hsa:5150 PDE7A, HCP1, PDE7; phosphodiesterase 7A (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=456
Score = 44.7 bits (104), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 30/105 (28%), Positives = 50/105 (47%), Gaps = 3/105 (2%)
Query 115 KIGTDWSFDCLHFGEISR-KPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVT-DCYKE 172
K+G +W+FD F ++ L + F + S L L L FL + D + +
Sbjct 124 KVG-NWNFDIFLFDRLTNGNSLVSLTFHLFSLHGLIEYFHLDMMKLRRFLVMIQEDYHSQ 182
Query 173 NPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
NPYHNA+H A V C L+ ++ ++ + + + IA+ HD
Sbjct 183 NPYHNAVHAADVTQAMHCYLKEPKLANSVTPWDILLSLIAAATHD 227
> mmu:29863 Pde7b; phosphodiesterase 7B (EC:3.1.4.17); K01120
3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=446
Score = 44.7 bits (104), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 33/112 (29%), Positives = 54/112 (48%), Gaps = 15/112 (13%)
Query 114 AKIGTDWSFDCLHFGEISRKP-----LCEI--GFAILSPFSLSPEISLHRDVLTAFLEEV 166
+K+GT W FD F ++ LC + ++ F L ++LHR FL V
Sbjct 110 SKVGT-WDFDIFLFDRLTNGNSLVTLLCHLFNSHGLIHHFKLD-MVTLHR-----FLVMV 162
Query 167 T-DCYKENPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
D + NPYHNA+H A V C L+ ++ +++ L+ + +A+ HD
Sbjct 163 QEDYHGHNPYHNAVHAADVTQAMHCYLKEPKLASFLTPLDIMLGLLAAAAHD 214
Lambda K H
0.324 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7009334380
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40