bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0785_orf2
Length=150
Score E
Sequences producing significant alignments: (Bits) Value
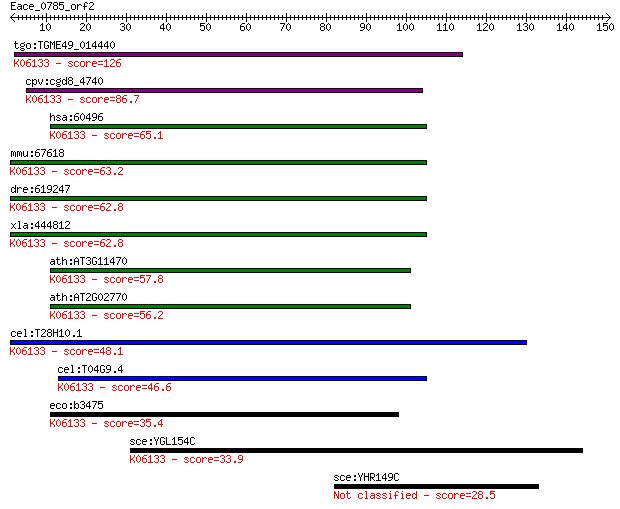
tgo:TGME49_014440 4'-phosphopantetheinyl transferase, putative... 126 3e-29
cpv:cgd8_4740 phosphopantetheinyl transferase ; K06133 4'-phos... 86.7 3e-17
hsa:60496 AASDHPPT, AASD-PPT, DKFZp566E2346, LYS2, LYS5; amino... 65.1 7e-11
mmu:67618 Aasdhppt, 2010309J24Rik, 2810407B07Rik, AASD-PPT, CG... 63.2 3e-10
dre:619247 MGC114148; zgc:114148; K06133 4'-phosphopantetheiny... 62.8 4e-10
xla:444812 aasdhppt, MGC84206; aminoadipate-semialdehyde dehyd... 62.8 4e-10
ath:AT3G11470 4'-phosphopantetheinyl transferase family protei... 57.8 1e-08
ath:AT2G02770 holo-[acyl-carrier-protein] synthase/ magnesium ... 56.2 4e-08
cel:T28H10.1 hypothetical protein; K06133 4'-phosphopantethein... 48.1 1e-05
cel:T04G9.4 hypothetical protein; K06133 4'-phosphopantetheiny... 46.6 3e-05
eco:b3475 acpT, ECK3459, JW3440, yhhU; holo-(acyl carrier prot... 35.4 0.060
sce:YGL154C LYS5; Lys5p (EC:1.2.1.31); K06133 4'-phosphopantet... 33.9 0.21
sce:YHR149C SKG6; Integral membrane protein that localizes pri... 28.5 7.8
> tgo:TGME49_014440 4'-phosphopantetheinyl transferase, putative
(EC:2.7.8.7); K06133 4'-phosphopantetheinyl transferase [EC:2.7.8.-]
Length=437
Score = 126 bits (316), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 61/117 (52%), Positives = 78/117 (66%), Gaps = 5/117 (4%)
Query 2 PSLGETAEFLHFNVSHDEGLVVFGASRHLVGVDCMRCSPRG--SRDTTQFLQQMKAHCTA 59
P E + F+HFNVSHD GLVV ASR LVGVDCM+ + RG SR L+ M + C
Sbjct 136 PHAEERSPFIHFNVSHDTGLVVLSASRLLVGVDCMQVTVRGASSRSVADLLRAMDSQCVG 195
Query 60 RDWAYIMGVRLP---EQQIRRFMRVWTVKEAFVKAIGTGIYIDPERLECTFPASGSP 113
R+ Y++G E+++RRFM++WT+KEAFVKA GTG+Y+DP RLEC P SP
Sbjct 196 RERPYVLGEDSDVSDEEKLRRFMKIWTMKEAFVKACGTGVYVDPARLECVLPEYDSP 252
> cpv:cgd8_4740 phosphopantetheinyl transferase ; K06133 4'-phosphopantetheinyl
transferase [EC:2.7.8.-]
Length=267
Score = 86.7 bits (213), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 43/108 (39%), Positives = 59/108 (54%), Gaps = 15/108 (13%)
Query 5 GETAEFLHFNVSHDEGLVVFGASRHLVGVDCMRCSPRGSR---------DTTQFLQQMKA 55
E+ LHFNVSHD +VV S+++VG+D M+ SR +FL MK
Sbjct 50 SESDSLLHFNVSHDGDVVVIILSKYMVGIDIMKTELSPSRVQLSNNIMEANEKFLNNMKN 109
Query 56 HCTARDWAYIMGVRLPEQQIRRFMRVWTVKEAFVKAIGTGIYIDPERL 103
+W YI ++ I +FM WT+KE+FVK IG G+Y+DP RL
Sbjct 110 VFHPSEWEYI------QKDISKFMHYWTIKESFVKYIGLGLYVDPRRL 151
> hsa:60496 AASDHPPT, AASD-PPT, DKFZp566E2346, LYS2, LYS5; aminoadipate-semialdehyde
dehydrogenase-phosphopantetheinyl transferase
(EC:2.7.8.-); K06133 4'-phosphopantetheinyl transferase
[EC:2.7.8.-]
Length=309
Score = 65.1 bits (157), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 34/95 (35%), Positives = 49/95 (51%), Gaps = 1/95 (1%)
Query 11 LHFNVSHDEGLVVFGASRHL-VGVDCMRCSPRGSRDTTQFLQQMKAHCTARDWAYIMGVR 69
+FN+SH V A L VG+D M+ S G +F MK T ++W I +
Sbjct 105 FNFNISHQGDYAVLAAEPELQVGIDIMKTSFPGRGSIPEFFHIMKRKFTNKEWETIRSFK 164
Query 70 LPEQQIRRFMRVWTVKEAFVKAIGTGIYIDPERLE 104
Q+ F R W +KE+F+KAIG G+ + +RLE
Sbjct 165 DEWTQLDMFYRNWALKESFIKAIGVGLGFELQRLE 199
> mmu:67618 Aasdhppt, 2010309J24Rik, 2810407B07Rik, AASD-PPT,
CGI-80, LYS2, LYS5; aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl
transferase (EC:2.7.8.-); K06133 4'-phosphopantetheinyl
transferase [EC:2.7.8.-]
Length=309
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 37/105 (35%), Positives = 52/105 (49%), Gaps = 2/105 (1%)
Query 1 KPSLGETAEFLHFNVSHDEGLVVFGASRHL-VGVDCMRCSPRGSRDTTQFLQQMKAHCTA 59
K SL F +FN+SH V A + VG+D M+ S G +F MK T
Sbjct 96 KDSLNPYPNF-NFNISHQGDYAVLAAEPEVQVGIDIMKTSFPGRGSIPEFFHIMKRKFTK 154
Query 60 RDWAYIMGVRLPEQQIRRFMRVWTVKEAFVKAIGTGIYIDPERLE 104
++W I Q+ F R W +KE+F+KAIG G+ + +RLE
Sbjct 155 KEWETIRSFNDEWTQLDMFYRHWALKESFIKAIGVGLGFEMQRLE 199
> dre:619247 MGC114148; zgc:114148; K06133 4'-phosphopantetheinyl
transferase [EC:2.7.8.-]
Length=293
Score = 62.8 bits (151), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 40/107 (37%), Positives = 52/107 (48%), Gaps = 3/107 (2%)
Query 1 KPSLGET--AEFLHFNVSHDEGLVVFGA-SRHLVGVDCMRCSPRGSRDTTQFLQQMKAHC 57
KP L +T A FNVSH V A + VG+D M+ S GS +F + M
Sbjct 78 KPYLPQTSSAPSWSFNVSHQGDYAVLAAEAGRQVGIDVMKTSRPGSSSVQEFFRIMNRQF 137
Query 58 TARDWAYIMGVRLPEQQIRRFMRVWTVKEAFVKAIGTGIYIDPERLE 104
T +W I Q+ F R W +KE+F KAIGTG+ D +R E
Sbjct 138 TDLEWTNIRTAGSDWDQLHMFYRHWALKESFNKAIGTGLGFDLQRAE 184
> xla:444812 aasdhppt, MGC84206; aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl
transferase; K06133 4'-phosphopantetheinyl
transferase [EC:2.7.8.-]
Length=302
Score = 62.8 bits (151), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 37/109 (33%), Positives = 56/109 (51%), Gaps = 5/109 (4%)
Query 1 KPSL--GETAEF--LHFNVSHDEGLVVFGAS-RHLVGVDCMRCSPRGSRDTTQFLQQMKA 55
KP L G ++E+ +FNVSH V A VGVD M+ GS +F + M
Sbjct 88 KPFLTGGSSSEYPCFNFNVSHQGDYAVLAAEPDRQVGVDIMKTDLPGSSSIEEFFRLMNR 147
Query 56 HCTARDWAYIMGVRLPEQQIRRFMRVWTVKEAFVKAIGTGIYIDPERLE 104
T ++W I + ++ F R W +KE+F+KAIG G+ + +R+E
Sbjct 148 QFTEKEWNSIRSMNNDWARLDMFYRHWALKESFIKAIGVGLGFNLQRIE 196
> ath:AT3G11470 4'-phosphopantetheinyl transferase family protein;
K06133 4'-phosphopantetheinyl transferase [EC:2.7.8.-]
Length=231
Score = 57.8 bits (138), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 30/91 (32%), Positives = 51/91 (56%), Gaps = 4/91 (4%)
Query 11 LHFNVSHDEGLVVFGASRHL-VGVDCMRCSPRGSRDTTQFLQQMKAHCTARDWAYIMGVR 69
LHFN+SH + L+ G + H+ VG+D + D F ++ +A + ++ +
Sbjct 55 LHFNISHTDSLIACGVTVHVPVGIDVEDKERKIKHDILAFAERF---YSADEVKFLSTLP 111
Query 70 LPEQQIRRFMRVWTVKEAFVKAIGTGIYIDP 100
PE Q + F+++WT+KEA+VKA+G G P
Sbjct 112 DPEVQRKEFIKLWTLKEAYVKALGKGFSAAP 142
> ath:AT2G02770 holo-[acyl-carrier-protein] synthase/ magnesium
ion binding; K06133 4'-phosphopantetheinyl transferase [EC:2.7.8.-]
Length=661
Score = 56.2 bits (134), Expect = 4e-08, Method: Composition-based stats.
Identities = 29/91 (31%), Positives = 50/91 (54%), Gaps = 4/91 (4%)
Query 11 LHFNVSHDEGLVVFGASRHL-VGVDCMRCSPRGSRDTTQFLQQMKAHCTARDWAYIMGVR 69
LHFN+SH + L+ G + H+ VG+D + D ++ +A + ++ +
Sbjct 429 LHFNISHTDSLISCGVTVHVPVGIDLEEMERKIKHDVLALAERFY---SADEVKFLSAIP 485
Query 70 LPEQQIRRFMRVWTVKEAFVKAIGTGIYIDP 100
PE Q + F+++WT+KEA+VKA+G G P
Sbjct 486 DPEVQRKEFIKLWTLKEAYVKALGKGFSGAP 516
> cel:T28H10.1 hypothetical protein; K06133 4'-phosphopantetheinyl
transferase [EC:2.7.8.-]
Length=299
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 39/137 (28%), Positives = 61/137 (44%), Gaps = 11/137 (8%)
Query 1 KPSLGETA-----EFLHFNVSHDEGLVVFGASRHLVGVDCMRCSPRGSRDTTQFLQQMKA 55
KPSL + + +NVSH LVV +GVD MR + ++ + +K
Sbjct 80 KPSLIQNKSDYSRQNFEYNVSHHGDLVVLATGDTRIGVDVMRVNEARRETASEQMNTLKR 139
Query 56 HCTARDWAYIMGVRLPE-QQIRRFMRVWTVKEAFVKAIGTGIYIDPERLEC-TFPASGS- 112
H + + + G E ++ F R+W +KE+ +KA G G+ P+ L TF S
Sbjct 140 HFSENEIEMVKGGDKCELKRWHAFYRIWCLKESILKATGVGL---PDGLHNHTFQVDSSY 196
Query 113 PQLSLDSIPQTDFHFRL 129
S S T ++ RL
Sbjct 197 DHASGHSTVSTQYYHRL 213
> cel:T04G9.4 hypothetical protein; K06133 4'-phosphopantetheinyl
transferase [EC:2.7.8.-]
Length=297
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 43/94 (45%), Gaps = 2/94 (2%)
Query 13 FNVSHDEGLVVFGAS-RHLVGVDCMRC-SPRGSRDTTQFLQQMKAHCTARDWAYIMGVRL 70
NVSH V F +S VGVD MR + R ++ +++ M + + +
Sbjct 100 LNVSHQGDYVAFASSCSSKVGVDVMRLDNERNNKTADEYINSMAKSASPEELRMMRSQPT 159
Query 71 PEQQIRRFMRVWTVKEAFVKAIGTGIYIDPERLE 104
++ F R W +KEA +KA G GI D L+
Sbjct 160 EAMKMTMFYRYWCLKEAILKATGVGIMKDLNSLD 193
> eco:b3475 acpT, ECK3459, JW3440, yhhU; holo-(acyl carrier protein)
synthase 2 (EC:2.7.8.7); K06133 4'-phosphopantetheinyl
transferase [EC:2.7.8.-]
Length=195
Score = 35.4 bits (80), Expect = 0.060, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 38/89 (42%), Gaps = 9/89 (10%)
Query 11 LHFNVSHD-EGLVVFGASRHLVGVDCMRCSPRGS-RDTTQFLQQMKAHCTARDWAYIMGV 68
L FN+SH + + + + VG D PR + R + + H M
Sbjct 67 LWFNLSHSGDDIALLLSDEGEVGCDIEVIRPRANWRWLANAVFSLGEHAE-------MDA 119
Query 69 RLPEQQIRRFMRVWTVKEAFVKAIGTGIY 97
P+QQ+ F R+WT KEA VK G +
Sbjct 120 VHPDQQLEMFWRIWTRKEAIVKQRGGSAW 148
> sce:YGL154C LYS5; Lys5p (EC:1.2.1.31); K06133 4'-phosphopantetheinyl
transferase [EC:2.7.8.-]
Length=272
Score = 33.9 bits (76), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 27/118 (22%), Positives = 55/118 (46%), Gaps = 16/118 (13%)
Query 31 VGVDCMRCSPRGSRDTTQFLQQMKAHCTARDWAYIMGVRLPEQQIRRFMRVWTVKEAFVK 90
VG+D G R+ + +++ + R++ ++ P F +W++KE++ K
Sbjct 133 VGIDIASPCNYGGREELELFKEVFSE---REFNGLLKASDP---CTIFTYLWSLKESYTK 186
Query 91 AIGTGIYIDPERLE----CTFPASGSPQ-LSLDSIPQTDFHFRLEEEEAPDYVICVCV 143
GTG+ D ++ FPA G+ ++LD +P FH + E ++ +C+
Sbjct 187 FTGTGLNTDLSLIDFGAISFFPAEGASMCITLDEVPLI-FHSQWFNNE----IVTICM 239
> sce:YHR149C SKG6; Integral membrane protein that localizes primarily
to growing sites such as the bud tip or the cell periphery;
potential Cdc28p substrate; Skg6p interacts with Zds1p
and Zds2p
Length=734
Score = 28.5 bits (62), Expect = 7.8, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 25/51 (49%), Gaps = 9/51 (17%)
Query 82 WTVKEAFVKAIGTGIYIDPERLECTFPASGSPQLSLDSIPQTDFHFRLEEE 132
+ V+ A AIG Y+DP +L PQ S DS DF R++E+
Sbjct 163 YNVRSATPPAIGRSWYVDPFQL---------PQESNDSNSLRDFAMRVQED 204
Lambda K H
0.324 0.139 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3134054208
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40