bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0483_orf2
Length=192
Score E
Sequences producing significant alignments: (Bits) Value
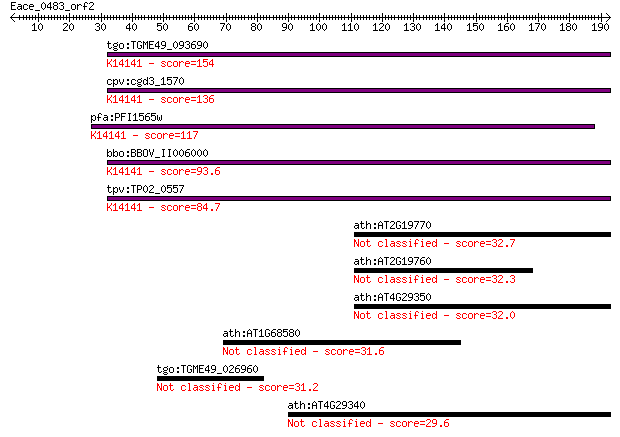
tgo:TGME49_093690 profilin family protein ; K14141 profilin-li... 154 2e-37
cpv:cgd3_1570 sporozoite antigen ; K14141 profilin-like protein 136 6e-32
pfa:PFI1565w PfPfn; profilin, putative; K14141 profilin-like p... 117 2e-26
bbo:BBOV_II006000 18.m06498; hypothetical protein; K14141 prof... 93.6 3e-19
tpv:TP02_0557 hypothetical protein; K14141 profilin-like protein 84.7 2e-16
ath:AT2G19770 PRF5; PRF5 (PROFILIN5); actin binding / actin mo... 32.7 0.84
ath:AT2G19760 PRF1; PRF1 (PROFILIN 1); actin binding 32.3 0.92
ath:AT4G29350 PFN2; PFN2 (PROFILIN 2); actin binding / protein... 32.0 1.4
ath:AT1G68580 agenet domain-containing protein / bromo-adjacen... 31.6 1.9
tgo:TGME49_026960 phosphofructokinase, putative (EC:2.7.1.90) 31.2 2.3
ath:AT4G29340 PRF4; PRF4 (PROFILIN 4); actin binding 29.6 6.3
> tgo:TGME49_093690 profilin family protein ; K14141 profilin-like
protein
Length=163
Score = 154 bits (388), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 82/162 (50%), Positives = 114/162 (70%), Gaps = 3/162 (1%)
Query 32 WDTSVKEWLVDTGKVYAGGIASIADGCRLFGAAIDNGEDAWSQLVKTGYQIEVLQEDGSS 91
WD VKEWLVDTG AGGIA+ DG AA + +D WS+L K ++ + + EDG++
Sbjct 4 WDPVVKEWLVDTGYCCAGGIANAEDGVVF--AAAADDDDGWSKLYKDDHEEDTIGEDGNA 61
Query 92 TQE-DCDEAETLRQAIVDGRAPNGVYIGGVKYKLAEVKRDFTYNDQNYDVAILGKNKGGG 150
+ +EA T++ A+ DG APNGV+IGG KYK+ ++ F YND +D+ + ++KGG
Sbjct 62 CGKVSINEASTIKAAVDDGSAPNGVWIGGQKYKVVRPEKGFEYNDCTFDITMCARSKGGA 121
Query 151 FLIKTPNDNVVIALYDEEKEQNKADALTTALAFAEYLYQGGF 192
LIKTPN ++VIALYDEEKEQ+K ++ T+ALAFAEYL+Q G+
Sbjct 122 HLIKTPNGSIVIALYDEEKEQDKGNSRTSALAFAEYLHQSGY 163
> cpv:cgd3_1570 sporozoite antigen ; K14141 profilin-like protein
Length=162
Score = 136 bits (342), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 75/162 (46%), Positives = 101/162 (62%), Gaps = 4/162 (2%)
Query 32 WDTSVKEWLVDTGKVYAGGIASIADGCRLFGAAIDNGEDAWSQLVKTGYQIEVLQEDG-S 90
WD VKEWL+DTG V AGG+ SI DG + A+ D G DAW LV+ ++ V+Q DG S
Sbjct 4 WDDMVKEWLIDTGSVCAGGLCSI-DGA-FYAASADQG-DAWKTLVREDHEENVIQSDGVS 60
Query 91 STQEDCDEAETLRQAIVDGRAPNGVYIGGVKYKLAEVKRDFTYNDQNYDVAILGKNKGGG 150
E ++ TL QAI +G+APNGV++GG KYK+ V++DF ND V + +GG
Sbjct 61 EAAELINDQTTLCQAISEGKAPNGVWVGGNKYKIIRVEKDFQQNDATVHVTFCNRPQGGC 120
Query 151 FLIKTPNDNVVIALYDEEKEQNKADALTTALAFAEYLYQGGF 192
FL+ T N VV+A+YDE K+Q+ + AL AEYL G+
Sbjct 121 FLVDTQNGTVVVAVYDESKDQSSGNCKKVALQLAEYLVSQGY 162
> pfa:PFI1565w PfPfn; profilin, putative; K14141 profilin-like
protein
Length=171
Score = 117 bits (293), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 63/165 (38%), Positives = 95/165 (57%), Gaps = 4/165 (2%)
Query 27 ADTQAWDTSVKEWLVDTGKVYAGGIASIADG----CRLFGAAIDNGEDAWSQLVKTGYQI 82
A+ +WD+ + + L+ T +V G+AS DG C G D D WS K Y I
Sbjct 2 AEEYSWDSYLNDRLLATNQVSGAGLASEEDGVVYACVAQGEESDPNFDKWSLFYKEDYDI 61
Query 83 EVLQEDGSSTQEDCDEAETLRQAIVDGRAPNGVYIGGVKYKLAEVKRDFTYNDQNYDVAI 142
EV E+G+ T + +E +T+ +G AP+GV++GG KY+ ++RD + N+DVA
Sbjct 62 EVEDENGTKTTKTINEGQTILVVFNEGYAPDGVWLGGTKYQFINIERDLEFEGYNFDVAT 121
Query 143 LGKNKGGGFLIKTPNDNVVIALYDEEKEQNKADALTTALAFAEYL 187
K KGG L+K P N+++ LYDEEKEQ++ ++ AL FA+ L
Sbjct 122 CAKLKGGLHLVKVPGGNILVVLYDEEKEQDRGNSKIAALTFAKEL 166
> bbo:BBOV_II006000 18.m06498; hypothetical protein; K14141 profilin-like
protein
Length=164
Score = 93.6 bits (231), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 53/162 (32%), Positives = 79/162 (48%), Gaps = 2/162 (1%)
Query 32 WDTSVKEWLVDTGKVYAGGIASIADGCRLFGAA-IDNGEDAWSQLVKTGYQIEVLQEDGS 90
W ++K+ + Y GIA+ DG LF AA ID+ + W + + Y+ E E+G
Sbjct 4 WVPTIKQLALADNACYGCGIANAEDG-ELFSAADIDHDDLCWDSVYRDPYEFEATDENGQ 62
Query 91 STQEDCDEAETLRQAIVDGRAPNGVYIGGVKYKLAEVKRDFTYNDQNYDVAILGKNKGGG 150
+ E T+ + R+ G++IGG KY A D D + KNKGG
Sbjct 63 PIKHQITEKATIMEVFEKRRSSIGIFIGGNKYTFANYDDDCPVGDYTFKCVSAAKNKGGA 122
Query 151 FLIKTPNDNVVIALYDEEKEQNKADALTTALAFAEYLYQGGF 192
L+KTP +VI ++DE + QNK + A A AEY+ G+
Sbjct 123 HLVKTPGGYIVICVFDENRGQNKTASRMAAFALAEYMAANGY 164
> tpv:TP02_0557 hypothetical protein; K14141 profilin-like protein
Length=164
Score = 84.7 bits (208), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 48/161 (29%), Positives = 75/161 (46%), Gaps = 0/161 (0%)
Query 32 WDTSVKEWLVDTGKVYAGGIASIADGCRLFGAAIDNGEDAWSQLVKTGYQIEVLQEDGSS 91
W +KE + Y GIA+ DG + +D+ W + K Y E + G+
Sbjct 4 WVPVLKETALSNNSCYGAGIANGEDGELFVASDLDHEGLHWDSVYKDPYVYETFDDAGNP 63
Query 92 TQEDCDEAETLRQAIVDGRAPNGVYIGGVKYKLAEVKRDFTYNDQNYDVAILGKNKGGGF 151
+ + DE T+R+ + G+++GG KY A D ++ KNKGG
Sbjct 64 LKINVDEKFTIREVFEKKMSSEGIFLGGEKYTFASYDPDMESGSFKFECVCGAKNKGGCH 123
Query 152 LIKTPNDNVVIALYDEEKEQNKADALTTALAFAEYLYQGGF 192
LIKTP + +V+ +YDE + Q+K + A AEYL G+
Sbjct 124 LIKTPGNYIVVVVYDETRGQDKTVSRMAAFNLAEYLATNGY 164
> ath:AT2G19770 PRF5; PRF5 (PROFILIN5); actin binding / actin
monomer binding
Length=134
Score = 32.7 bits (73), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 21/82 (25%), Positives = 37/82 (45%), Gaps = 11/82 (13%)
Query 111 APNGVYIGGVKYKLAEVKRDFTYNDQNYDVAILGKNKGGGFLIKTPNDNVVIALYDEEKE 170
AP G+++ G+KY + + + + I GK GG IK ++V LY+E
Sbjct 64 APTGMFLAGLKYMVIQGEPN---------AVIRGKKGAGGITIKKTGQSMVFGLYEEPVT 114
Query 171 QNKADALTTALAFAEYLYQGGF 192
+ + + L +YL + G
Sbjct 115 PGQCNMVVERL--GDYLIEQGL 134
> ath:AT2G19760 PRF1; PRF1 (PROFILIN 1); actin binding
Length=131
Score = 32.3 bits (72), Expect = 0.92, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 27/57 (47%), Gaps = 9/57 (15%)
Query 111 APNGVYIGGVKYKLAEVKRDFTYNDQNYDVAILGKNKGGGFLIKTPNDNVVIALYDE 167
AP G+++GG KY + + ++ I GK GG IK N +V YDE
Sbjct 61 APTGLFLGGEKYMVIQGEQG---------AVIRGKKGPGGVTIKKTNQALVFGFYDE 108
> ath:AT4G29350 PFN2; PFN2 (PROFILIN 2); actin binding / protein
binding
Length=131
Score = 32.0 bits (71), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 22/82 (26%), Positives = 35/82 (42%), Gaps = 11/82 (13%)
Query 111 APNGVYIGGVKYKLAEVKRDFTYNDQNYDVAILGKNKGGGFLIKTPNDNVVIALYDEEKE 170
AP G+++GG KY + + + I GK GG IK +V +YDE
Sbjct 61 APTGLFLGGEKYMVVQGEAG---------AVIRGKKGPGGVTIKKTTQALVFGIYDEPMT 111
Query 171 QNKADALTTALAFAEYLYQGGF 192
+ + + L +YL + G
Sbjct 112 GGQCNLVVERL--GDYLIESGL 131
> ath:AT1G68580 agenet domain-containing protein / bromo-adjacent
homology (BAH) domain-containing protein
Length=648
Score = 31.6 bits (70), Expect = 1.9, Method: Composition-based stats.
Identities = 25/99 (25%), Positives = 39/99 (39%), Gaps = 23/99 (23%)
Query 69 EDAWSQLVKTGYQIEVLQEDG-----------------------SSTQEDCDEAETLRQA 105
ED S +K G IEVL ED Q+ DE++ L +
Sbjct 356 EDGSSHHIKKGSLIEVLSEDSGIRGCWFKALVLKKHKDKVKVQYQDIQDADDESKKLEEW 415
Query 106 IVDGRAPNGVYIGGVKYKLAEVKRDFTYNDQNYDVAILG 144
I+ R G ++G ++ K +V R + DV ++G
Sbjct 416 ILTSRVAAGDHLGDLRIKGRKVVRPMLKPSKENDVCVIG 454
> tgo:TGME49_026960 phosphofructokinase, putative (EC:2.7.1.90)
Length=1225
Score = 31.2 bits (69), Expect = 2.3, Method: Composition-based stats.
Identities = 16/34 (47%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 48 AGGIASIADGCRLFGAAIDNGEDAWSQLVKTGYQ 81
A G+ S+A+G LF + EDA SQLV G Q
Sbjct 1141 ANGVFSVAEGDALFALSESASEDAVSQLVAQGMQ 1174
> ath:AT4G29340 PRF4; PRF4 (PROFILIN 4); actin binding
Length=134
Score = 29.6 bits (65), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 25/103 (24%), Positives = 40/103 (38%), Gaps = 19/103 (18%)
Query 90 SSTQEDCDEAETLRQAIVDGRAPNGVYIGGVKYKLAEVKRDFTYNDQNYDVAILGKNKGG 149
S +D DE L AP G+++ G KY + + + I GK G
Sbjct 51 SDIMKDFDEPGHL--------APTGLFMAGAKYMVIQGEPG---------AVIRGKKGAG 93
Query 150 GFLIKTPNDNVVIALYDEEKEQNKADALTTALAFAEYLYQGGF 192
G IK + V +Y+E + + + L +YL + G
Sbjct 94 GITIKKTGQSCVFGIYEEPVTPGQCNMVVERL--GDYLLEQGL 134
Lambda K H
0.317 0.136 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5558593832
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40