bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0473_orf1
Length=123
Score E
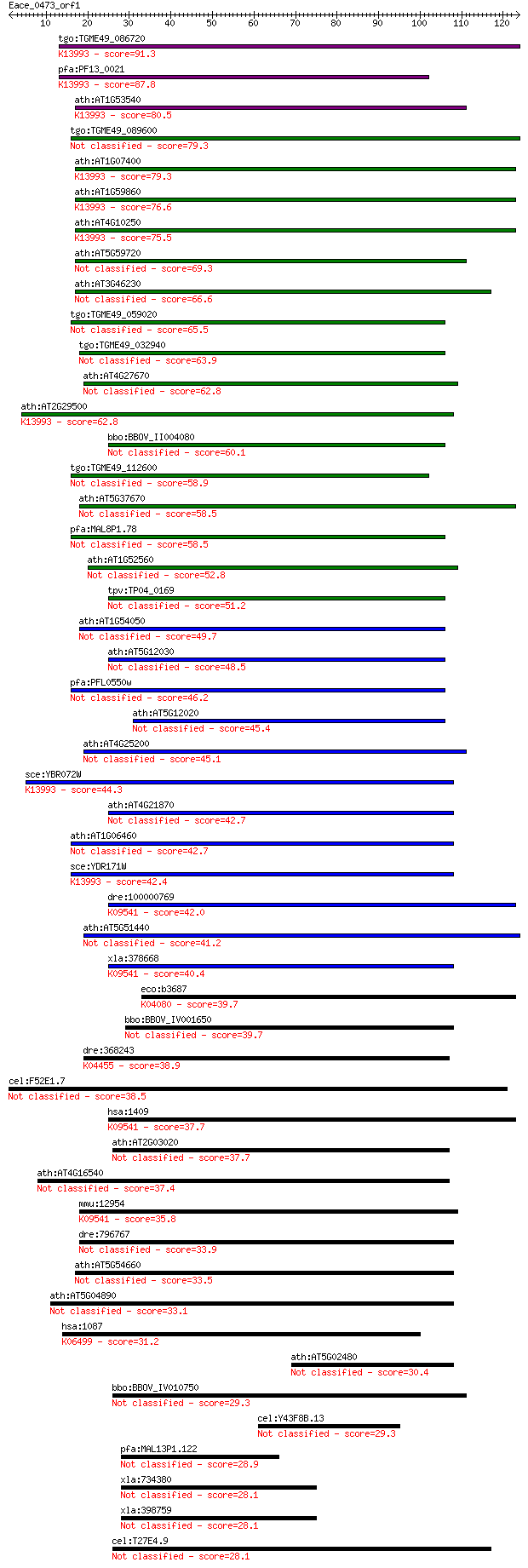
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_086720 heat shock protein 28 ; K13993 HSP20 family ... 91.3 8e-19
pfa:PF13_0021 small heat shock protein, putative; K13993 HSP20... 87.8 7e-18
ath:AT1G53540 17.6 kDa class I small heat shock protein (HSP17... 80.5 1e-15
tgo:TGME49_089600 Hsp20/alpha crystallin domain-containing pro... 79.3 2e-15
ath:AT1G07400 17.8 kDa class I heat shock protein (HSP17.8-CI)... 79.3 3e-15
ath:AT1G59860 17.6 kDa class I heat shock protein (HSP17.6A-CI... 76.6 2e-14
ath:AT4G10250 ATHSP22.0; K13993 HSP20 family protein 75.5 4e-14
ath:AT5G59720 HSP18.2 (heat shock protein 18.2) 69.3 2e-12
ath:AT3G46230 ATHSP17.4 66.6 2e-11
tgo:TGME49_059020 bradyzoite antigen, putative 65.5 4e-11
tgo:TGME49_032940 small heat shock protein 20 63.9
ath:AT4G27670 HSP21; HSP21 (HEAT SHOCK PROTEIN 21) 62.8 3e-10
ath:AT2G29500 17.6 kDa class I small heat shock protein (HSP17... 62.8 3e-10
bbo:BBOV_II004080 18.m06338; small heat shock protein 60.1 1e-09
tgo:TGME49_112600 small heat shock protein 21 (EC:1.1.2.4) 58.9 3e-09
ath:AT5G37670 15.7 kDa class I-related small heat shock protei... 58.5 4e-09
pfa:MAL8P1.78 small heat shock protein, putative 58.5 5e-09
ath:AT1G52560 26.5 kDa class I small heat shock protein-like (... 52.8 2e-07
tpv:TP04_0169 small heat shock protein 51.2 8e-07
ath:AT1G54050 17.4 kDa class III heat shock protein (HSP17.4-C... 49.7 2e-06
ath:AT5G12030 AT-HSP17.6A (ARABIDOPSIS THALIANA HEAT SHOCK PRO... 48.5 5e-06
pfa:PFL0550w HSP20-like chaperone 46.2 3e-05
ath:AT5G12020 HSP17.6II (17.6 KDA CLASS II HEAT SHOCK PROTEIN) 45.4 4e-05
ath:AT4G25200 ATHSP23.6-MITO (MITOCHONDRION-LOCALIZED SMALL HE... 45.1 6e-05
sce:YBR072W HSP26; Hsp26p; K13993 HSP20 family protein 44.3 1e-04
ath:AT4G21870 26.5 kDa class P-related heat shock protein (HSP... 42.7 2e-04
ath:AT1G06460 ACD32.1 (ALPHA-CRYSTALLIN DOMAIN 32.1) 42.7 3e-04
sce:YDR171W HSP42; Hsp42p; K13993 HSP20 family protein 42.4 3e-04
dre:100000769 cryaa, MGC92036, fc06h04, wu:fc06h04, zgc:92036;... 42.0 5e-04
ath:AT5G51440 23.5 kDa mitochondrial small heat shock protein ... 41.2 8e-04
xla:378668 cryaa, cryaa-A; crystallin, alpha A; K09541 crystal... 40.4 0.001
eco:b3687 ibpA, ECK3679, hslT, htpN, JW3664; heat shock chaper... 39.7 0.002
bbo:BBOV_IV001650 21.m02864; hypothetical protein 39.7 0.002
dre:368243 hspb1, cb153, hsp1, hsp25, hsp27, zgc:103437; heat ... 38.9 0.004
cel:F52E1.7 hsp-17; Heat Shock Protein family member (hsp-17) 38.5 0.005
hsa:1409 CRYAA, CRYA1, HSPB4; crystallin, alpha A; K09541 crys... 37.7 0.008
ath:AT2G03020 heat shock protein-related 37.7 0.008
ath:AT4G16540 heat shock protein-related 37.4 0.010
mmu:12954 Cryaa, AI323437, Acry-1, Crya-1, Crya1, DAcry-1, lop... 35.8 0.032
dre:796767 hspb11, cb02d06, mg:cb02d06; heat shock protein, al... 33.9 0.14
ath:AT5G54660 heat shock protein-related 33.5 0.18
ath:AT5G04890 RTM2; RTM2 (RESTRICTED TEV MOVEMENT 2) 33.1 0.20
hsa:1087 CEACAM7, CEA, CGM2; carcinoembryonic antigen-related ... 31.2 0.92
ath:AT5G02480 hypothetical protein 30.4 1.5
bbo:BBOV_IV010750 23.m06090; hypothetical protein 29.3 3.2
cel:Y43F8B.13 hypothetical protein 29.3 3.5
pfa:MAL13P1.122 SET domain protein, putative 28.9 3.8
xla:734380 atad3a-a, MGC85169, atad3, atad3-a; ATPase family, ... 28.1 6.7
xla:398759 atad3a-b, MGC68616, atad3, atad3-b; ATPase family, ... 28.1 7.0
cel:T27E4.9 hsp-16.49; Heat Shock Protein family member (hsp-1... 28.1 7.6
> tgo:TGME49_086720 heat shock protein 28 ; K13993 HSP20 family
protein
Length=276
Score = 91.3 bits (225), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 49/114 (42%), Positives = 79/114 (69%), Gaps = 7/114 (6%)
Query 13 SCIPRIDLKDAGDKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSNWYMQER 72
+P++D+KD G + V+HAD+PG+D+++++V+ HDG L I ++ ++ +Y+QER
Sbjct 167 GSMPKVDMKDTGSEFVVHADVPGMDRENLRVDVHDGVLRISGTQRDEKQQQEEGFYLQER 226
Query 73 CSSNFYRSFQLPANIQEEAIKATYNNGVLEVNIPK-TPQVQTTQPP--RSINVE 123
S+F RSF LP ++E+ IKA+ NGVL+V++PK TP TQPP R+I +E
Sbjct 227 SQSSFSRSFVLPDKVKEDQIKASLTNGVLQVHVPKETP----TQPPAIRNITIE 276
> pfa:PF13_0021 small heat shock protein, putative; K13993 HSP20
family protein
Length=211
Score = 87.8 bits (216), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 41/91 (45%), Positives = 60/91 (65%), Gaps = 2/91 (2%)
Query 13 SCIPRIDLKDAGDKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDS--NWYMQ 70
S +P +D+ D L + D+PG++K+DV++ DG+L I KS E KD +Y++
Sbjct 101 SNVPAMDVLDKEKHLEIKMDVPGLNKEDVQINLDDGKLEISGEFKKSHEQKDEQQRYYIK 160
Query 71 ERCSSNFYRSFQLPANIQEEAIKATYNNGVL 101
ERC S+FYRSF LP N+ E+ IKAT+ +GVL
Sbjct 161 ERCESSFYRSFTLPENVSEDEIKATFKDGVL 191
> ath:AT1G53540 17.6 kDa class I small heat shock protein (HSP17.6C-CI)
(AA 1-156); K13993 HSP20 family protein
Length=157
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 39/96 (40%), Positives = 61/96 (63%), Gaps = 3/96 (3%)
Query 17 RIDLKDAGDKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSN--WYMQERCS 74
++D ++ + V AD+PG+ K++VKVE DG ++Q + +S E+++ N W+ ER S
Sbjct 51 KVDWRETPEAHVFKADLPGLRKEEVKVEVEDGN-ILQISGERSNENEEKNDKWHRVERSS 109
Query 75 SNFYRSFQLPANIQEEAIKATYNNGVLEVNIPKTPQ 110
F R F+LP N + E IKA+ NGVL V +PK P+
Sbjct 110 GKFTRRFRLPENAKMEEIKASMENGVLSVTVPKVPE 145
> tgo:TGME49_089600 Hsp20/alpha crystallin domain-containing protein
Length=272
Score = 79.3 bits (194), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 50/113 (44%), Positives = 67/113 (59%), Gaps = 7/113 (6%)
Query 16 PRID-LKDAG-DKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHK---DSNWYMQ 70
PR+D DA ++LVL D+PG +KKDV++E G L I K EE K D N ++
Sbjct 162 PRLDAYYDANRNRLVLLFDLPGFEKKDVEIELDKGALAISGERPKLEESKLGQDCNNIIK 221
Query 71 ERCSSNFYRSFQLPANIQEEAIKATYNNGVLEVNIPKTPQVQTTQPPRSINVE 123
ER FYR FQLP N +EE+IKA+ GVLEV I + Q T+ + I+V+
Sbjct 222 ERSFGFFYRKFQLPGNAEEESIKASMEQGVLEVTIGLKDENQPTK--KKIDVQ 272
> ath:AT1G07400 17.8 kDa class I heat shock protein (HSP17.8-CI);
K13993 HSP20 family protein
Length=157
Score = 79.3 bits (194), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 38/107 (35%), Positives = 62/107 (57%), Gaps = 1/107 (0%)
Query 17 RIDLKDAGDKLVLHADIPGIDKKDVKVEAHDGRLV-IQAASNKSEEHKDSNWYMQERCSS 75
R+D K+ + V AD+PG+ K++VKVE D ++ I + +E K W+ ER S
Sbjct 49 RVDWKETAEAHVFKADLPGMKKEEVKVEIEDDSVLKISGERHVEKEEKQDTWHRVERSSG 108
Query 76 NFYRSFQLPANIQEEAIKATYNNGVLEVNIPKTPQVQTTQPPRSINV 122
F R F+LP N++ + +KA+ NGVL V +PK + + +SI++
Sbjct 109 QFSRKFKLPENVKMDQVKASMENGVLTVTVPKVEEAKKKAQVKSIDI 155
> ath:AT1G59860 17.6 kDa class I heat shock protein (HSP17.6A-CI);
K13993 HSP20 family protein
Length=155
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 38/107 (35%), Positives = 61/107 (57%), Gaps = 1/107 (0%)
Query 17 RIDLKDAGDKLVLHADIPGIDKKDVKVEAHDGRLV-IQAASNKSEEHKDSNWYMQERCSS 75
R+D K+ + V AD+PG+ K++VKVE D ++ I + +E K W+ ER S
Sbjct 47 RVDWKETAEAHVFKADLPGMKKEEVKVEIEDDSVLKISGERHVEKEEKQDTWHRVERSSG 106
Query 76 NFYRSFQLPANIQEEAIKATYNNGVLEVNIPKTPQVQTTQPPRSINV 122
F R F+LP N++ + +KA+ NGVL V +PK + +SI++
Sbjct 107 GFSRKFRLPENVKMDQVKASMENGVLTVTVPKVETNKKKAQVKSIDI 153
> ath:AT4G10250 ATHSP22.0; K13993 HSP20 family protein
Length=195
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 37/107 (34%), Positives = 59/107 (55%), Gaps = 2/107 (1%)
Query 17 RIDLKDAGDKLVLHADIPGIDKKDVKVEAHD-GRLVIQAASNKSEEHKDSNWYMQERCSS 75
R+D K+ + + DIPG+ K +VK+E + G L + + EE K W+ ER
Sbjct 72 RVDWKETAEGHEIMLDIPGLKKDEVKIEVEENGVLRVSGERKREEEKKGDQWHRVERSYG 131
Query 76 NFYRSFQLPANIQEEAIKATYNNGVLEVNIPKTPQVQTTQPPRSINV 122
F+R F+LP N+ E++KA NGVL +N+ K + + PR +N+
Sbjct 132 KFWRQFKLPDNVDMESVKAKLENGVLTINLTKLSP-EKVKGPRVVNI 177
> ath:AT5G59720 HSP18.2 (heat shock protein 18.2)
Length=161
Score = 69.3 bits (168), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 40/95 (42%), Positives = 57/95 (60%), Gaps = 1/95 (1%)
Query 17 RIDLKDAGDKLVLHADIPGIDKKDVKVEAHDGRLV-IQAASNKSEEHKDSNWYMQERCSS 75
R+D K+ + V AD+PG+ K++VKVE D ++ I +K E K+ W+ ER S
Sbjct 53 RVDWKETPEAHVFKADLPGLKKEEVKVEVEDKNVLQISGERSKENEEKNDKWHRVERASG 112
Query 76 NFYRSFQLPANIQEEAIKATYNNGVLEVNIPKTPQ 110
F R F+LP N + E +KAT NGVL V +PK P+
Sbjct 113 KFMRRFRLPENAKMEEVKATMENGVLTVVVPKAPE 147
> ath:AT3G46230 ATHSP17.4
Length=156
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 40/101 (39%), Positives = 60/101 (59%), Gaps = 4/101 (3%)
Query 17 RIDLKDAGDKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSE-EHKDSNWYMQERCSS 75
++D ++ + V AD+PG+ K++VKVE DG ++ + SE E K W+ ER S
Sbjct 50 KVDWRETPEAHVFKADVPGLKKEEVKVEVEDGNILQISGERSSENEEKSDTWHRVERSSG 109
Query 76 NFYRSFQLPANIQEEAIKATYNNGVLEVNIPKTPQVQTTQP 116
F R F+LP N + E +KA+ NGVL V +PK VQ ++P
Sbjct 110 KFMRRFRLPENAKVEEVKASMENGVLSVTVPK---VQESKP 147
> tgo:TGME49_059020 bradyzoite antigen, putative
Length=229
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 35/96 (36%), Positives = 53/96 (55%), Gaps = 6/96 (6%)
Query 16 PRIDLKDAGDK--LVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSNW----YM 69
PR+D++ K +++ AD+PG+ K DV +E +G +VI+ E K + +
Sbjct 119 PRVDVEFDSKKKEMIILADLPGLQKDDVTIEVDNGAIVIKGEKTSKEAEKVDDGKTKNIL 178
Query 70 QERCSSNFYRSFQLPANIQEEAIKATYNNGVLEVNI 105
ER S F R FQLP+N + + I A +NGVL V I
Sbjct 179 TERVSGYFARRFQLPSNYKPDGISAAMDNGVLRVTI 214
> tgo:TGME49_032940 small heat shock protein 20
Length=192
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 37/92 (40%), Positives = 54/92 (58%), Gaps = 6/92 (6%)
Query 18 IDLKDAGDKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSE--EHKDSNWYMQ--ERC 73
D K++ LV+ D+PG K DV VE +G+L I +K+E E +N + ER
Sbjct 76 FDKKESSIDLVM--DLPGFTKDDVSVEVGEGQLFISGPRSKNELREKYGANLVLSIHERP 133
Query 74 SSNFYRSFQLPANIQEEAIKATYNNGVLEVNI 105
+ F+RSFQLP N E++++A NG+LEV I
Sbjct 134 TGFFFRSFQLPPNAVEDSVRAVMTNGLLEVKI 165
> ath:AT4G27670 HSP21; HSP21 (HEAT SHOCK PROTEIN 21)
Length=227
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 34/90 (37%), Positives = 51/90 (56%), Gaps = 4/90 (4%)
Query 19 DLKDAGDKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSNWYMQERCSSNFY 78
D+K+ ++ + D+PG+ K+DVK+ D LVI+ K E D +W R S++
Sbjct 130 DIKEEEHEIKMRFDMPGLSKEDVKISVEDNVLVIKGEQKK--EDSDDSW--SGRSVSSYG 185
Query 79 RSFQLPANIQEEAIKATYNNGVLEVNIPKT 108
QLP N +++ IKA NGVL + IPKT
Sbjct 186 TRLQLPDNCEKDKIKAELKNGVLFITIPKT 215
> ath:AT2G29500 17.6 kDa class I small heat shock protein (HSP17.6B-CI);
K13993 HSP20 family protein
Length=153
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 41/107 (38%), Positives = 61/107 (57%), Gaps = 3/107 (2%)
Query 4 SSASWFTHNSCI--PRIDLKDAGDKLVLHADIPGIDKKDVKVEAH-DGRLVIQAASNKSE 60
+S+S NS I R+D ++ + V AD+PG+ K++VKVE D L I + +
Sbjct 32 TSSSLSRENSAIVNARVDWRETPEAHVFKADLPGLKKEEVKVEIEEDSVLKISGERHVEK 91
Query 61 EHKDSNWYMQERCSSNFYRSFQLPANIQEEAIKATYNNGVLEVNIPK 107
E K+ W+ ER S F R F+LP N++ + +KA NGVL V +PK
Sbjct 92 EDKNDTWHRVERSSGQFTRRFRLPENVKMDQVKAAMENGVLTVTVPK 138
> bbo:BBOV_II004080 18.m06338; small heat shock protein
Length=188
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 30/85 (35%), Positives = 50/85 (58%), Gaps = 4/85 (4%)
Query 25 DKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHK----DSNWYMQERCSSNFYRS 80
+KLVL ++PG D+ VE G L+I NK E ++ + + +++ER FYR
Sbjct 87 NKLVLLMELPGFSSTDINVECGWGELIISGPRNKDELYEKFGNNLDIHIRERKVGYFYRR 146
Query 81 FQLPANIQEEAIKATYNNGVLEVNI 105
F+LP N +++I Y+NG+L++ I
Sbjct 147 FKLPNNAIDKSISVGYSNGILDIRI 171
> tgo:TGME49_112600 small heat shock protein 21 (EC:1.1.2.4)
Length=195
Score = 58.9 bits (141), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 32/92 (34%), Positives = 49/92 (53%), Gaps = 6/92 (6%)
Query 16 PRID--LKDAGDKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEH----KDSNWYM 69
PR+D A K+V+ D+PG +KKD+ VE D ++I ++ + + S +
Sbjct 83 PRLDAYYDTAAHKIVMLFDLPGFEKKDISVEVTDHAIIISGTRDRLNDKVLYGEKSRELI 142
Query 70 QERCSSNFYRSFQLPANIQEEAIKATYNNGVL 101
+ER +F R FQLP N E+A+ A GVL
Sbjct 143 KERAFGHFCRKFQLPTNAVEDAVTAAMTGGVL 174
> ath:AT5G37670 15.7 kDa class I-related small heat shock protein-like
(HSP15.7-CI)
Length=137
Score = 58.5 bits (140), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 32/111 (28%), Positives = 64/111 (57%), Gaps = 7/111 (6%)
Query 18 IDLKDAGDKLVLHADIPGIDKKDVKVEAHDGRLV-IQAASNKSEEHKDSNWYMQER---- 72
ID ++ + + ++PG +K+D+KV+ +G ++ I+ K E+ ++ W++ ER
Sbjct 24 IDWMESNNSHIFKINVPGYNKEDIKVQIEEGNVLSIRGEGIKEEKKENLVWHVAEREAFS 83
Query 73 -CSSNFYRSFQLPANIQEEAIKATYNNGVLEVNIPKTPQVQTTQPPRSINV 122
S F R +LP N++ + +KA NGVL V +PK ++++ R++N+
Sbjct 84 GGGSEFLRRIELPENVKVDQVKAYVENGVLTVVVPKDTSSKSSK-VRNVNI 133
> pfa:MAL8P1.78 small heat shock protein, putative
Length=172
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 34/96 (35%), Positives = 54/96 (56%), Gaps = 6/96 (6%)
Query 16 PRIDLKDAGDKL--VLHADIPGIDKKDVKVEAHDGRLVIQAASNKSE---EHKDS-NWYM 69
P++D+ DK +L DIPG +D+ VE +G L + +++E + DS +
Sbjct 61 PKVDIMYDADKCEAILVLDIPGFKIEDIDVEIGEGMLTVAGPRSQTELFETYGDSLVLHA 120
Query 70 QERCSSNFYRSFQLPANIQEEAIKATYNNGVLEVNI 105
+ER F R F+LP NI ++ KATY NG+LE+ +
Sbjct 121 KEREVGYFKRIFKLPNNILDDTAKATYKNGILEIKM 156
> ath:AT1G52560 26.5 kDa class I small heat shock protein-like
(HSP26.5-P)
Length=225
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 33/92 (35%), Positives = 51/92 (55%), Gaps = 4/92 (4%)
Query 20 LKDAGDKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDS---NWYMQERCSSN 76
+K+ D L ++PG+ K+DVK+ +DG L I+ +K+EE K S + Y +
Sbjct 122 VKEQDDCYKLRYEVPGLTKEDVKITVNDGILTIK-GDHKAEEEKGSPEEDEYWSSKSYGY 180
Query 77 FYRSFQLPANIQEEAIKATYNNGVLEVNIPKT 108
+ S LP + + E IKA NGVL + IP+T
Sbjct 181 YNTSLSLPDDAKVEDIKAELKNGVLNLVIPRT 212
> tpv:TP04_0169 small heat shock protein
Length=203
Score = 51.2 bits (121), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 25/85 (29%), Positives = 44/85 (51%), Gaps = 4/85 (4%)
Query 25 DKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHK----DSNWYMQERCSSNFYRS 80
+ LV ++PG D+++ DG +++ K E ++ + N ++ER FYR
Sbjct 76 NNLVTLMELPGFPISDIEINIGDGEMIVSGPRPKEELYEKFGENLNIQIRERKVGYFYRR 135
Query 81 FQLPANIQEEAIKATYNNGVLEVNI 105
F+LP N + A Y+NG+L + I
Sbjct 136 FKLPNNAMDNTASACYDNGILSIII 160
> ath:AT1G54050 17.4 kDa class III heat shock protein (HSP17.4-CIII)
Length=155
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 52/94 (55%), Gaps = 6/94 (6%)
Query 18 IDLKDAGDKLVLHADIPGIDKKDVKVEAHDGR-LVIQAASNKS---EEHKDSNWY--MQE 71
ID+ ++ + + + DIPGI K D++V + R LVI++ + +E ++ + Y ++
Sbjct 44 IDILESPKEYIFYLDIPGISKSDIQVTVEEERTLVIKSNGKRKRDDDESEEGSKYIRLER 103
Query 72 RCSSNFYRSFQLPANIQEEAIKATYNNGVLEVNI 105
R + N + F+LP + ++ A Y GVL V I
Sbjct 104 RLAQNLVKKFRLPEDADMASVTAKYQEGVLTVVI 137
> ath:AT5G12030 AT-HSP17.6A (ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN
17.6A); unfolded protein binding
Length=156
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 44/83 (53%), Gaps = 2/83 (2%)
Query 25 DKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNK--SEEHKDSNWYMQERCSSNFYRSFQ 82
D V D+PGI +++V+ + +++ + + ++E++ + ER F R FQ
Sbjct 55 DAYVFAVDMPGIKGDEIQVQIENENVLVVSGKRQRDNKENEGVKFVRMERRMGKFMRKFQ 114
Query 83 LPANIQEEAIKATYNNGVLEVNI 105
LP N E I A N+GVL+V I
Sbjct 115 LPDNADLEKISAACNDGVLKVTI 137
> pfa:PFL0550w HSP20-like chaperone
Length=231
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 33/97 (34%), Positives = 49/97 (50%), Gaps = 7/97 (7%)
Query 16 PRIDLKDAGDKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKS--EEHKDSNWY---MQ 70
P+I++ D VL DIPG+ K+++KVE G L + K EE + N Y +
Sbjct 119 PKIEIYSTCDFAVLMMDIPGVSKENLKVELEKGLLKVYGNKYKPHIEELEKRNEYHTKII 178
Query 71 ERCSSNFY-RSFQLPANIQE-EAIKATYNNGVLEVNI 105
ER + ++ + FQ+P E + I NNG L V I
Sbjct 179 ERLNEYYFCKIFQMPPAFSEGQNISCKLNNGELLVKI 215
> ath:AT5G12020 HSP17.6II (17.6 KDA CLASS II HEAT SHOCK PROTEIN)
Length=155
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 41/77 (53%), Gaps = 2/77 (2%)
Query 31 ADIPGIDKKDVKVEAHDGRLVIQAASNKSE--EHKDSNWYMQERCSSNFYRSFQLPANIQ 88
D+PGI ++KV+ + +++ + + E E++ + ER F R FQLP N
Sbjct 60 VDMPGIKGDEIKVQVENDNVLVVSGERQRENKENEGVKYVRMERRMGKFMRKFQLPENAD 119
Query 89 EEAIKATYNNGVLEVNI 105
+ I A ++GVL+V +
Sbjct 120 LDKISAVCHDGVLKVTV 136
> ath:AT4G25200 ATHSP23.6-MITO (MITOCHONDRION-LOCALIZED SMALL
HEAT SHOCK PROTEIN 23.6)
Length=210
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 31/93 (33%), Positives = 46/93 (49%), Gaps = 5/93 (5%)
Query 19 DLKDAGDKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSNWYMQERCSSNFY 78
D+K+ D L L D+PG+ ++DVK+ LVI+ E+ + R +S
Sbjct 110 DIKEKDDALYLRIDMPGLSREDVKLALEQDTLVIRGEGKNEEDGGEEGESGNRRFTSR-- 167
Query 79 RSFQLPANIQE-EAIKATYNNGVLEVNIPKTPQ 110
LP I + + IKA NGVL+V IPK +
Sbjct 168 --IGLPDKIYKIDEIKAEMKNGVLKVVIPKMKE 198
> sce:YBR072W HSP26; Hsp26p; K13993 HSP20 family protein
Length=214
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 35/110 (31%), Positives = 53/110 (48%), Gaps = 9/110 (8%)
Query 5 SASWFTHNSCIPRIDLKDAGDKLVLHADIPGI-DKKDVKVEAHDGRLVI----QAASNKS 59
S F + +P +D+ D + L +PG+ KKD+ +E H + I + S +
Sbjct 83 SGFGFPRSVAVP-VDILDHDNNYELKVVVPGVKSKKDIDIEYHQNKNQILVSGEIPSTLN 141
Query 60 EEHKDSNWYMQERCSSNFYRSFQLP--ANIQEEAIKATYNNGVLEVNIPK 107
EE KD ++E S F R LP + + IKA Y NGVL + +PK
Sbjct 142 EESKDK-VKVKESSSGKFKRVITLPDYPGVDADNIKADYANGVLTLTVPK 190
> ath:AT4G21870 26.5 kDa class P-related heat shock protein (HSP26.5-P)
Length=134
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 37/83 (44%), Gaps = 7/83 (8%)
Query 25 DKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSNWYMQERCSSNFYRSFQLP 84
D D+PG+ K+++KVE D +I D + F R F+LP
Sbjct 35 DSHTFSVDLPGLRKEEIKVEIEDSIYLIIRTEATPMSPPD-------QPLKTFKRKFRLP 87
Query 85 ANIQEEAIKATYNNGVLEVNIPK 107
+I I A Y +GVL V +PK
Sbjct 88 ESIDMIGISAGYEDGVLTVIVPK 110
> ath:AT1G06460 ACD32.1 (ALPHA-CRYSTALLIN DOMAIN 32.1)
Length=285
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 23/99 (23%), Positives = 48/99 (48%), Gaps = 7/99 (7%)
Query 16 PRIDLKDAGDKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSN-------WY 68
PR ++ ++ V+ ++PG D++VE + L + + D+ ++
Sbjct 186 PRSNVAESTHSYVVAIELPGASINDIRVEVDNTNLTVTGRRTSICQKVDAGTKASILGYH 245
Query 69 MQERCSSNFYRSFQLPANIQEEAIKATYNNGVLEVNIPK 107
QE F S+ LP+N+ ++ + A + +G+L + IPK
Sbjct 246 KQEILQGPFKVSWPLPSNVNKDNVSAEFMDGILRIVIPK 284
> sce:YDR171W HSP42; Hsp42p; K13993 HSP20 family protein
Length=375
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/96 (26%), Positives = 47/96 (48%), Gaps = 4/96 (4%)
Query 16 PRIDLKDAGDKLVLHADIPGIDKKDVKVEAHDG--RLVIQAASNKSEEHKDSNWYMQERC 73
P +++ D D V+ +PG + + ++ H ++I+ + + E
Sbjct 244 PEVNVYDTEDTYVVVLALPGANSRAFHIDYHPSSHEMLIKGKIEDRVGIDEKFLKITELK 303
Query 74 SSNFYRSFQLPA--NIQEEAIKATYNNGVLEVNIPK 107
F R+ + P I++E IKATYNNG+L++ +PK
Sbjct 304 YGAFERTVKFPVLPRIKDEEIKATYNNGLLQIKVPK 339
> dre:100000769 cryaa, MGC92036, fc06h04, wu:fc06h04, zgc:92036;
crystallin, alpha A; K09541 crystallin, alpha A
Length=173
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 51/99 (51%), Gaps = 8/99 (8%)
Query 25 DKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSNWYMQERCSSNFYRSFQLP 84
+K ++ D+ ++ V+ D + IQ K E +D + Y+ S F+R ++LP
Sbjct 70 EKFTVYLDVKHFSPDELSVKVTDDYVEIQG---KHGERQDDHGYI----SREFHRRYRLP 122
Query 85 ANIQEEAIKATYN-NGVLEVNIPKTPQVQTTQPPRSINV 122
+N+ + AI T + +G+L + PKT + + R+I V
Sbjct 123 SNVDQSAITCTLSADGLLTLCGPKTSGIDAGRGDRTIPV 161
> ath:AT5G51440 23.5 kDa mitochondrial small heat shock protein
(HSP23.5-M)
Length=210
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 33/106 (31%), Positives = 50/106 (47%), Gaps = 8/106 (7%)
Query 19 DLKDAGDKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSNWYMQERCSSNFY 78
++K+ D L L D+PG+ ++DVK+ LVI+ E S F
Sbjct 112 NVKEKDDALHLRIDMPGLSREDVKLALEQNTLVIRGEGETEEGEDVSG------DGRRFT 165
Query 79 RSFQLPANI-QEEAIKATYNNGVLEVNIPKTPQVQTTQPPRSINVE 123
+LP + + + IKA NGVL+V IPK + + R INV+
Sbjct 166 SRIELPEKVYKTDEIKAEMKNGVLKVVIPKIKEDERNN-IRHINVD 210
> xla:378668 cryaa, cryaa-A; crystallin, alpha A; K09541 crystallin,
alpha A
Length=171
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/84 (27%), Positives = 46/84 (54%), Gaps = 8/84 (9%)
Query 25 DKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSNWYMQERCSSNFYRSFQLP 84
D+ V++ D+ +D+ V+ HD + I N E +D + Y+ S F+R ++LP
Sbjct 69 DRFVINLDVKHFSPEDLSVKVHDDFVEIHGKHN---ERQDDHGYI----SREFHRRYRLP 121
Query 85 ANIQEEAIKATYN-NGVLEVNIPK 107
+N+ + ++ T + +G+L + PK
Sbjct 122 SNMDQNSVSCTLSADGILTLFGPK 145
> eco:b3687 ibpA, ECK3679, hslT, htpN, JW3664; heat shock chaperone;
K04080 molecular chaperone IbpA
Length=137
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/90 (26%), Positives = 44/90 (48%), Gaps = 6/90 (6%)
Query 33 IPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSNWYMQERCSSNFYRSFQLPANIQEEAI 92
+ G + ++++ A D LV++ A ++E K+ + Q NF R FQL NI
Sbjct 53 VAGFAESELEITAQDNLLVVKGAH--ADEQKERTYLYQGIAERNFERKFQLAENIHVRG- 109
Query 93 KATYNNGVLEVNIPKTPQVQTTQPPRSINV 122
A NG+L +++ + + + PR I +
Sbjct 110 -ANLVNGLLYIDLERV--IPEAKKPRRIEI 136
> bbo:BBOV_IV001650 21.m02864; hypothetical protein
Length=288
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 38/79 (48%), Gaps = 3/79 (3%)
Query 29 LHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSNWYMQERCSSNFYRSFQLPANIQ 88
L IP ++ K++K++ DG L + S +E K + + R S +F S LP +
Sbjct 211 LSFTIPNVNAKNIKIKIRDGLLTV---SGHAERVKRGDHESEMRYSESFQHSVSLPRDAM 267
Query 89 EEAIKATYNNGVLEVNIPK 107
E KA N L + IPK
Sbjct 268 ELDAKARLRNDTLTIEIPK 286
> dre:368243 hspb1, cb153, hsp1, hsp25, hsp27, zgc:103437; heat
shock protein, alpha-crystallin-related, 1; K04455 heat shock
27kDa protein 1
Length=199
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 27/89 (30%), Positives = 43/89 (48%), Gaps = 8/89 (8%)
Query 19 DLKDAGDKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSNWYMQERCSSNFY 78
++K GD + D+ +++ V+ DG L I + K EE KD + ++ RC F
Sbjct 94 EVKQTGDSWKISLDVNHFSPEELNVKTKDGVLEI---TGKHEERKDEHGFIS-RC---FT 146
Query 79 RSFQLPANIQEEAIKATYN-NGVLEVNIP 106
R + LP + E I + + GVL V P
Sbjct 147 RKYTLPPGVDSEKISSCLSPEGVLTVEAP 175
> cel:F52E1.7 hsp-17; Heat Shock Protein family member (hsp-17)
Length=148
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 30/121 (24%), Positives = 56/121 (46%), Gaps = 8/121 (6%)
Query 1 PHGSSASWFTHNSCIPRIDLKDAGDKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSE 60
P+ + + T + ID+ + + + D+ + +++KV D +L+I+ N+
Sbjct 30 PYWADQTMLTGHRVGDAIDVVNNDQEYNVSVDVSQFEPEELKVNIVDNQLIIEGKHNEK- 88
Query 61 EHKDSNWYMQERCSSNFYRSFQLPANIQEEAIKATY-NNGVLEVNIPKTPQVQTTQPPRS 119
++ Y Q +F R + LP ++ E IK+ NNGVL V K + Q P +
Sbjct 89 ----TDKYGQ--VERHFVRKYNLPTGVRPEQIKSELSNNGVLTVKYEKNQEQQPKSIPIT 142
Query 120 I 120
I
Sbjct 143 I 143
> hsa:1409 CRYAA, CRYA1, HSPB4; crystallin, alpha A; K09541 crystallin,
alpha A
Length=173
Score = 37.7 bits (86), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 49/100 (49%), Gaps = 9/100 (9%)
Query 25 DKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSNWYMQERCSSNFYRSFQLP 84
DK V+ D+ +D+ V+ D + I N E +D + Y+ S F+R ++LP
Sbjct 69 DKFVIFLDVKHFSPEDLTVKVQDDFVEIHGKHN---ERQDDHGYI----SREFHRRYRLP 121
Query 85 ANIQEEAIKATYN-NGVLEVNIPKTPQ-VQTTQPPRSINV 122
+N+ + A+ + + +G+L PK + T R+I V
Sbjct 122 SNVDQSALSCSLSADGMLTFCGPKIQTGLDATHAERAIPV 161
> ath:AT2G03020 heat shock protein-related
Length=255
Score = 37.7 bits (86), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 21/81 (25%), Positives = 34/81 (41%), Gaps = 6/81 (7%)
Query 26 KLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSNWYMQERCSSNFYRSFQLPA 85
KL + AD+PG+ K++ V +GR+ + + +Y S P
Sbjct 179 KLFVRADMPGVPKENFTVSVTNGRVKVTGQAPAVSHDSSGRFY------SGDVAMLSTPV 232
Query 86 NIQEEAIKATYNNGVLEVNIP 106
+I IK NGV+ + IP
Sbjct 233 DIPSRRIKTIAKNGVIRLLIP 253
> ath:AT4G16540 heat shock protein-related
Length=232
Score = 37.4 bits (85), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 21/99 (21%), Positives = 40/99 (40%), Gaps = 6/99 (6%)
Query 8 WFTHNSCIPRIDLKDAGDKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSNW 67
F + ++ +K KL + AD+PG+ K++ V +GR+ + + +
Sbjct 138 CFCQQFLVAQVSVKLPNGKLFVRADMPGVPKENFTVSVTNGRVKVTGEAPALSHDSSGRF 197
Query 68 YMQERCSSNFYRSFQLPANIQEEAIKATYNNGVLEVNIP 106
Y + P +I IK +GV+ + IP
Sbjct 198 YSGDGA------MLSTPVDIPSRRIKTIAKDGVIRLLIP 230
> mmu:12954 Cryaa, AI323437, Acry-1, Crya-1, Crya1, DAcry-1, lop18;
crystallin, alpha A; K09541 crystallin, alpha A
Length=196
Score = 35.8 bits (81), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 22/92 (23%), Positives = 47/92 (51%), Gaps = 8/92 (8%)
Query 18 IDLKDAGDKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSNWYMQERCSSNF 77
+ ++ DK V+ D+ +D+ V+ + + I N E +D + Y+ S F
Sbjct 85 VKVRSDRDKFVIFLDVKHFSPEDLTVKVLEDFVEIHGKHN---ERQDDHGYI----SREF 137
Query 78 YRSFQLPANIQEEAIKATYN-NGVLEVNIPKT 108
+R ++LP+N+ + A+ + + +G+L + PK
Sbjct 138 HRRYRLPSNVDQSALSCSLSADGMLTFSGPKV 169
> dre:796767 hspb11, cb02d06, mg:cb02d06; heat shock protein,
alpha-crystallin-related, b11
Length=205
Score = 33.9 bits (76), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 20/90 (22%), Positives = 37/90 (41%), Gaps = 3/90 (3%)
Query 18 IDLKDAGDKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSNWYMQERCSSNF 77
L G L D +++ V+ +L + + K ++ D RC F
Sbjct 78 FQLGKEGSHYALTLDTQDFSPEELAVKQVGRKLRVSGKTEKKQD--DGKGSYSYRCQE-F 134
Query 78 YRSFQLPANIQEEAIKATYNNGVLEVNIPK 107
+ F LP + E++ + NNG L++ P+
Sbjct 135 RQEFDLPEGVNPESVSCSLNNGQLQIQAPR 164
> ath:AT5G54660 heat shock protein-related
Length=192
Score = 33.5 bits (75), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 28/99 (28%), Positives = 47/99 (47%), Gaps = 9/99 (9%)
Query 17 RIDLKDAGDKLVLHADIPGIDKKDVKVEAH-DGRLV---IQAASNKSEEHKDSNWYMQER 72
R+D VL +DIP + K +V+V +GR++ Q SNK + +S+W
Sbjct 88 RVDWSQTDQAYVLKSDIPVVGKNNVQVYVDINGRVMEISGQWNSNK-KAATNSDWRSGRW 146
Query 73 CSSNFYRSFQLPANIQEEAIKATYNN----GVLEVNIPK 107
+ R +LP++ + +A +N LE+ IPK
Sbjct 147 WEHGYVRRLELPSDADAKYSEAFLSNNDDYSFLEIRIPK 185
> ath:AT5G04890 RTM2; RTM2 (RESTRICTED TEV MOVEMENT 2)
Length=366
Score = 33.1 bits (74), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 22/97 (22%), Positives = 42/97 (43%), Gaps = 9/97 (9%)
Query 11 HNSCIPRIDLKDAGDKLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSNWYMQ 70
+ +P+ + KD + +L+ D+ G K+ +KV +I+ + +
Sbjct 16 YEDFVPKSEWKDQPEATILNIDLTGFAKEQMKVTYVHSSKMIRVTGERP---------LA 66
Query 71 ERCSSNFYRSFQLPANIQEEAIKATYNNGVLEVNIPK 107
R S F F +P N + I ++ N VL + +PK
Sbjct 67 NRKWSRFNEVFTVPQNCLVDKIHGSFKNNVLTITMPK 103
> hsa:1087 CEACAM7, CEA, CGM2; carcinoembryonic antigen-related
cell adhesion molecule 7; K06499 carcinoembryonic antigen-related
cell adhesion molecule
Length=265
Score = 31.2 bits (69), Expect = 0.92, Method: Compositional matrix adjust.
Identities = 24/89 (26%), Positives = 37/89 (41%), Gaps = 5/89 (5%)
Query 14 CIPRIDLKDAGDKLVLHADIPGIDKKDVKV---EAHDGRLVIQAASNKSEEHKDSNWYMQ 70
CIP L L ++P + ++ V +G+ V+ N+S+ NWY
Sbjct 12 CIPWQGLLLTASLLTFW-NLPNSAQTNIDVVPFNVAEGKEVLLVVHNESQNLYGYNWYKG 70
Query 71 ERCSSNFYRSFQLPANIQEEAIKATYNNG 99
ER +N YR NI +E +NG
Sbjct 71 ERVHAN-YRIIGYVKNISQENAPGPAHNG 98
> ath:AT5G02480 hypothetical protein
Length=508
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 19/43 (44%), Positives = 22/43 (51%), Gaps = 4/43 (9%)
Query 69 MQERCS-SNFYRSFQLPANIQEEAIKATYNNG---VLEVNIPK 107
M E C F R QLP I EEA Y +G VLE+ +PK
Sbjct 445 MAEHCPPGEFMREIQLPNRIPEEANIEAYFDGTGPVLEIVVPK 487
> bbo:BBOV_IV010750 23.m06090; hypothetical protein
Length=1157
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 27/103 (26%), Positives = 40/103 (38%), Gaps = 18/103 (17%)
Query 26 KLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEE--------HKDSNWYMQERC---- 73
K +LH + + DVK H SNKS E H+D + +ER
Sbjct 1055 KRLLHQHVMNMLMDDVKNNIHRSPATTSTVSNKSHEEIERLQILHRDQIEFAEERVNQLS 1114
Query 74 ------SSNFYRSFQLPANIQEEAIKATYNNGVLEVNIPKTPQ 110
S ++Y + + N+Q+E A N L + KT Q
Sbjct 1115 SQVEVLSKSYYIAEEKAMNLQKELDNALEVNAQLNSLLEKTRQ 1157
> cel:Y43F8B.13 hypothetical protein
Length=465
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 11/34 (32%), Positives = 18/34 (52%), Gaps = 0/34 (0%)
Query 61 EHKDSNWYMQERCSSNFYRSFQLPANIQEEAIKA 94
E + W ERC NF+ LP +++E +K+
Sbjct 377 EKLSTAWDFAERCHKNFFNIVFLPVELRQEILKS 410
> pfa:MAL13P1.122 SET domain protein, putative
Length=2548
Score = 28.9 bits (63), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 28 VLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDS 65
+L DI GI KK++K+ +D ++ + + +E HKD+
Sbjct 1862 ILSNDIVGIMKKELKISLYDFIMLKKKTISTNENHKDN 1899
> xla:734380 atad3a-a, MGC85169, atad3, atad3-a; ATPase family,
AAA domain containing 3A
Length=593
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 12/47 (25%), Positives = 28/47 (59%), Gaps = 0/47 (0%)
Query 28 VLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSNWYMQERCS 74
++H D+PG+++++ V + + V+Q AS + K + + ++CS
Sbjct 466 IVHFDLPGLEERERLVRLYFDKYVLQPASEGKQRLKVAQFDYGKKCS 512
> xla:398759 atad3a-b, MGC68616, atad3, atad3-b; ATPase family,
AAA domain containing 3A
Length=593
Score = 28.1 bits (61), Expect = 7.0, Method: Composition-based stats.
Identities = 12/47 (25%), Positives = 28/47 (59%), Gaps = 0/47 (0%)
Query 28 VLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSNWYMQERCS 74
++H D+PG+++++ V + + V+Q AS + K + + ++CS
Sbjct 466 IVHFDLPGLEERERLVRLYFDKYVLQPASEGKQRLKVAQFDYGKKCS 512
> cel:T27E4.9 hsp-16.49; Heat Shock Protein family member (hsp-16.49)
Length=143
Score = 28.1 bits (61), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 22/92 (23%), Positives = 37/92 (40%), Gaps = 9/92 (9%)
Query 26 KLVLHADIPGIDKKDVKVEAHDGRLVIQAASNKSEEHKDSNWYMQERCSSNFYRSFQLPA 85
K + D+ +D+K+E L I+ K EH S +F + LP
Sbjct 53 KFSVQLDVSHFKPEDLKIELDGRELKIEGIQEKKSEHGYSK--------RSFSKMILLPE 104
Query 86 NIQEEAIK-ATYNNGVLEVNIPKTPQVQTTQP 116
++ ++K A N G L++ PK + P
Sbjct 105 DVDLTSVKSAISNEGKLQIEAPKKTNSSRSIP 136
Lambda K H
0.313 0.129 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2003197800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40