bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0346_orf2
Length=253
Score E
Sequences producing significant alignments: (Bits) Value
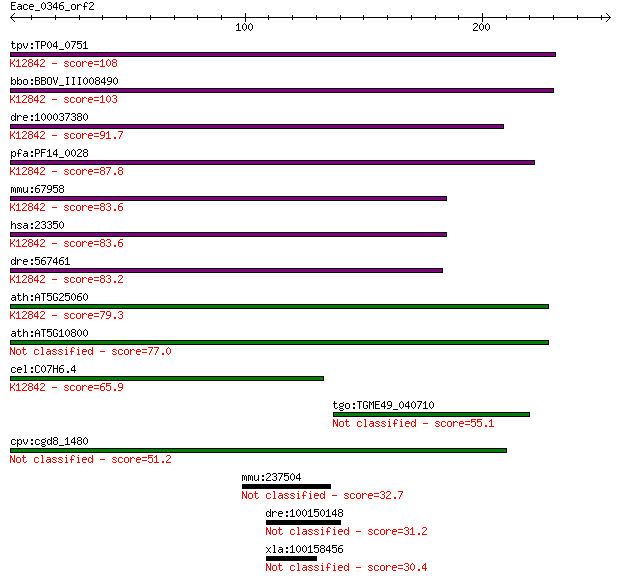
tpv:TP04_0751 hypothetical protein; K12842 U2-associated prote... 108 1e-23
bbo:BBOV_III008490 17.m07743; surp module family protein; K128... 103 7e-22
dre:100037380 zgc:163098; K12842 U2-associated protein SR140 91.7 2e-18
pfa:PF14_0028 pre-mRNA splicing factor, putative; K12842 U2-as... 87.8 4e-17
mmu:67958 2610101N10Rik, AU023006, Sr140, U2surp; RIKEN cDNA 2... 83.6 6e-16
hsa:23350 U2SURP, KIAA0332, MGC133197, SR140, fSAPa; U2 snRNP-... 83.6 6e-16
dre:567461 KIAA0332, mp:zf637-1-000918, wu:fa03e02, wu:fb38b01... 83.2 8e-16
ath:AT5G25060 RNA recognition motif (RRM)-containing protein; ... 79.3 1e-14
ath:AT5G10800 RNA recognition motif (RRM)-containing protein 77.0 5e-14
cel:C07H6.4 hypothetical protein; K12842 U2-associated protein... 65.9 1e-10
tgo:TGME49_040710 RRM domain-containing protein 55.1 2e-07
cpv:cgd8_1480 RPR domain containing protein, present in protei... 51.2 3e-06
mmu:237504 Rassf9, AW322107, MGC41694, Pamci; Ras association ... 32.7 1.4
dre:100150148 ras association domain-containing protein 9-like 31.2 3.4
xla:100158456 rassf9, P-CIP1; Ras association (RalGDS/AF-6) do... 30.4 6.9
> tpv:TP04_0751 hypothetical protein; K12842 U2-associated protein
SR140
Length=730
Score = 108 bits (271), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 66/232 (28%), Positives = 113/232 (48%), Gaps = 28/232 (12%)
Query 1 LLRNITRDRASICAAMSFCMSHGSSSAEVCSCLTESLTLPDTALTMRLARMYLLSDLLAN 60
+LR +T R IC AM F +++ S+ + L P+T + +++ +Y++SD+L N
Sbjct 408 ILRGVTSVRNDICNAMLFVINNSESAYHLTDLLFNYFNDPNTNVQQKISILYVISDVLYN 467
Query 61 SSAQTAHAWTYRHHLEKHLPFIADRWSKVLEDLRLGVGVGGASPRTQQEQWQGVERALYK 120
SS+ ++W YR+ +EKHLP + + E T + Q + A+ K
Sbjct 468 SSSSKQYSWVYRNSIEKHLPQLFHSIKQYKE------------KSTSKISSQQLIDAVMK 515
Query 121 MTKLWEAWGVYPPAFLRGIEATLCGRDITA--TILDPEAAAAAAAKCEAAALELADPQLD 178
+ +W++W VY FL G+EATL G D+ + T+ + E + + ++ D
Sbjct 516 LLSIWDSWTVYSQQFLNGLEATLLGDDLDSFKTLPEFEQHKDLLSTNDGVVVDYFD---- 571
Query 179 GEPLAENLRPLIAQFPLWLRPAAMEWLLLDRYQLDRLCQQRGLGVLPTRSSS 230
L+A PL R A ++LL+ +L +C QRGL V P+ +S
Sbjct 572 ----------LLATLPLKYRETAYKYLLMRLKELKSMCLQRGLLVQPSDRNS 613
> bbo:BBOV_III008490 17.m07743; surp module family protein; K12842
U2-associated protein SR140
Length=717
Score = 103 bits (256), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 71/230 (30%), Positives = 107/230 (46%), Gaps = 24/230 (10%)
Query 1 LLRNITRDRASICAAMSFCMSHGSSSAEVCSCLTESLTLPDTALTMRLARMYLLSDLLAN 60
+L N T R I AM F ++HG S+ +V CL E + + +++R+Y+LSD+L N
Sbjct 393 ILSNATTIRGYIADAMMFMINHGESAVQVTDCLVEYIMKDSPTVDTKISRLYILSDVLYN 452
Query 61 SSAQTAHAWTYRHHLEKHLPFIADRWSKVLEDLRLGVGVGGASPRTQQEQWQGVERALYK 120
+SA W YR EK +P + ++ + E ++ S QE VER L
Sbjct 453 TSASHQFGWIYRLTFEKKIPEV---FAHIREYIK-----SSTSKIAVQELISCVERIL-- 502
Query 121 MTKLWEAWGVYPPAFLRGIEATLCGRDITATILDPEAAAAAAAKCEAAALELADPQLDGE 180
W W YP +L G+E+ L G P + ++ DP DG+
Sbjct 503 --NAWHQWDAYPQEYLYGLESMLWG---------PYPSEFVTLPIFDQHRDIIDPGYDGD 551
Query 181 PLAENLRPLIAQFPLWLRPAAMEWLLLDRYQLDRLCQQRGLGVLP-TRSS 229
L L+++F L RP A E+ L+ +L +LCQ RG+ P TR S
Sbjct 552 EFI--LFDLLSRFTLKWRPVAYEYSLMRLKKLKQLCQNRGVVSTPYTRES 599
> dre:100037380 zgc:163098; K12842 U2-associated protein SR140
Length=874
Score = 91.7 bits (226), Expect = 2e-18, Method: Composition-based stats.
Identities = 64/210 (30%), Positives = 103/210 (49%), Gaps = 19/210 (9%)
Query 1 LLRNITRDRASICAAMSFCMSHGSSSAEVCSCLTESLTLPDTALTMRLARMYLLSDLLAN 60
LLR +T R I AM FC+ ++ E+ SC+TESL++ T L ++AR+YL+SD+L N
Sbjct 436 LLRGLTPRRDEIGDAMLFCLERAEAAEEIVSCITESLSIAHTPLQKKIARLYLISDILYN 495
Query 61 SSAQTAHAWTYRHHLEKHLPFIADRWSKVLEDLRLGVGVGGASPRTQQEQWQGVERALYK 120
S A+ A+A YR E LP I S+ +++ R Q EQ++ + +
Sbjct 496 SCAKVANASYYRKFFETKLPEIFGDISEAYRNIQ---------ARLQAEQFK---QRIMG 543
Query 121 MTKLWEAWGVYPPAFLRGIEATLCG--RDITATILDPEAAAAAAAKCEAAALELADPQLD 178
+ WE W VYP +FL ++ G + I PE + + L ++D
Sbjct 544 CFRAWEDWAVYPDSFLIQLQNIFLGLIKPGEEVIERPEPESPDLDGAPLDGVPLDRGEID 603
Query 179 GEPLAENLRPLIAQFPLWLRPAAMEWLLLD 208
G PL + + PL P+A++ + +D
Sbjct 604 GTPLDD-----LDGSPLTWDPSAIDGVPVD 628
> pfa:PF14_0028 pre-mRNA splicing factor, putative; K12842 U2-associated
protein SR140
Length=655
Score = 87.8 bits (216), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 54/227 (23%), Positives = 94/227 (41%), Gaps = 36/227 (15%)
Query 1 LLRNITRDRASICAAMSFCMSHGSSSAEVCSCLTESLT--LPDTALTMRLARMYLLSDLL 58
+L + + R SIC AM FC H S ++ ++ LT D +++ +YLLSD+L
Sbjct 403 ILSTLNKKRVSICRAMIFCTRHSDYSLDIIKIISNFLTDVKYDLLKKVKINLIYLLSDIL 462
Query 59 ANSSAQTAHAWTYRHHLEKHLP----FIADRWSKVLEDLRLGVGVGGASPRTQQEQWQGV 114
N S + +W+YR H+E LP F+ KV ++ + +
Sbjct 463 YNCSNEFFSSWSYRKHIEDELPRIFYFLRKHIKKVDSKIKGKLFID-------------- 508
Query 115 ERALYKMTKLWEAWGVYPPAFLRGIEATLCGRDITATILDPEAAAAAAAKCEAAALELAD 174
+L + +W W +Y F+ G+ LC I ++ + ++ D
Sbjct 509 --SLINIFNMWNCWAIYNSVFVNGLLCLLCNMKINYIDVNKSEEENDEENIDGHKIQYYD 566
Query 175 PQLDGEPLAENLRPLIAQFPLWLRPAAMEWLLLDRYQLDRLCQQRGL 221
I ++PL LR A + + ++++LC QRGL
Sbjct 567 D--------------IKRYPLSLRRNAYMYFKKNDDEINKLCLQRGL 599
> mmu:67958 2610101N10Rik, AU023006, Sr140, U2surp; RIKEN cDNA
2610101N10 gene; K12842 U2-associated protein SR140
Length=1029
Score = 83.6 bits (205), Expect = 6e-16, Method: Composition-based stats.
Identities = 58/186 (31%), Positives = 91/186 (48%), Gaps = 22/186 (11%)
Query 1 LLRNITRDRASICAAMSFCMSHGSSSAEVCSCLTESLTLPDTALTMRLARMYLLSDLLAN 60
+LR +T + I AM FC+++ ++ E+ C+TESL++ T L ++AR+YL+SD+L N
Sbjct 545 ILRGLTPRKNDIGDAMVFCLNNAEAAEEIVDCITESLSILKTPLPKKIARLYLVSDVLYN 604
Query 61 SSAQTAHAWTYRHHLEKHLPFIADRWSKVLEDLRLGVGVGGASPRTQQEQWQGVERALYK 120
SSA+ A+A YR E L ++ DL A+ RT Q Q
Sbjct 605 SSAKVANASYYRKFFETKL-------CQIFSDL-------NATYRTIQGHLQSENFKQRV 650
Query 121 MT--KLWEAWGVYPPAFLRGIEATLCGRDITATILDPEAAAAAAAKCEAAALELADPQLD 178
MT + WE W +YP FL ++ G I++ + + A +E +LD
Sbjct 651 MTCFRAWEDWAIYPEPFLIKLQNIFLG---LVNIIEEKETEDVPDDLDGAPIE---EELD 704
Query 179 GEPLAE 184
G PL +
Sbjct 705 GAPLED 710
> hsa:23350 U2SURP, KIAA0332, MGC133197, SR140, fSAPa; U2 snRNP-associated
SURP domain containing; K12842 U2-associated protein
SR140
Length=1029
Score = 83.6 bits (205), Expect = 6e-16, Method: Composition-based stats.
Identities = 58/186 (31%), Positives = 91/186 (48%), Gaps = 22/186 (11%)
Query 1 LLRNITRDRASICAAMSFCMSHGSSSAEVCSCLTESLTLPDTALTMRLARMYLLSDLLAN 60
+LR +T + I AM FC+++ ++ E+ C+TESL++ T L ++AR+YL+SD+L N
Sbjct 545 ILRGLTPRKNDIGDAMVFCLNNAEAAEEIVDCITESLSILKTPLPKKIARLYLVSDVLYN 604
Query 61 SSAQTAHAWTYRHHLEKHLPFIADRWSKVLEDLRLGVGVGGASPRTQQEQWQGVERALYK 120
SSA+ A+A YR E L ++ DL A+ RT Q Q
Sbjct 605 SSAKVANASYYRKFFETKL-------CQIFSDL-------NATYRTIQGHLQSENFKQRV 650
Query 121 MT--KLWEAWGVYPPAFLRGIEATLCGRDITATILDPEAAAAAAAKCEAAALELADPQLD 178
MT + WE W +YP FL ++ G I++ + + A +E +LD
Sbjct 651 MTCFRAWEDWAIYPEPFLIKLQNIFLG---LVNIIEEKETEDVPDDLDGAPIE---EELD 704
Query 179 GEPLAE 184
G PL +
Sbjct 705 GAPLED 710
> dre:567461 KIAA0332, mp:zf637-1-000918, wu:fa03e02, wu:fb38b01,
wu:fc75d03; si:dkey-181m9.9; K12842 U2-associated protein
SR140
Length=968
Score = 83.2 bits (204), Expect = 8e-16, Method: Composition-based stats.
Identities = 57/190 (30%), Positives = 90/190 (47%), Gaps = 26/190 (13%)
Query 1 LLRNITRDRASICAAMSFCMSHGSSSAEVCSCLTESLTLPDTALTMRLARMYLLSDLLAN 60
LLR +T ++ I M FC++H ++ E+ C+ ESL++ T L ++AR+YL+SD+L N
Sbjct 480 LLRGLTPRKSDIAGGMFFCLTHADAAEEIVECIAESLSILKTPLPKKIARLYLVSDVLYN 539
Query 61 SSAQTAHAWTYRHHLEKHLPFIADRWSKVLEDLRLGVGVGGASPRTQQE--QWQGVERAL 118
SSA+ +A YR E L ++ DL A+ +T Q Q + ++ +
Sbjct 540 SSAKVTNASYYRKFFETKL-------CQIFSDL-------NATYKTIQGHLQSENFKQRV 585
Query 119 YKMTKLWEAWGVYPPAFLRGIEATLCGRDITATILDPEAAAAAAAKCEAAALE------L 172
+ WE W VYP FL ++ G LDPE +E +
Sbjct 586 MSCFRAWEEWAVYPDPFLIKLQNIFLG----LVSLDPEKEPEELVPELPEKVEDIDGAPI 641
Query 173 ADPQLDGEPL 182
+ +LDG PL
Sbjct 642 VEEELDGAPL 651
> ath:AT5G25060 RNA recognition motif (RRM)-containing protein;
K12842 U2-associated protein SR140
Length=946
Score = 79.3 bits (194), Expect = 1e-14, Method: Composition-based stats.
Identities = 61/232 (26%), Positives = 113/232 (48%), Gaps = 30/232 (12%)
Query 1 LLRNITRDRASICAAMSFCMSHGSSSAEVCSCLTESLTLPDTALTMRLARMYLLSDLLAN 60
+LR +T +R+ I AM F + + ++ EV LTESLTL +T++ ++AR+ L+SD+L N
Sbjct 448 MLRALTLERSQIKEAMGFALDNADAAGEVVEVLTESLTLKETSIPTKVARLMLVSDILHN 507
Query 61 SSAQTAHAWTYRHHLEKHLPFIADRWSKVLEDLRLGVGVGGASPRTQQEQWQGVERALYK 120
SSA+ +A YR E LP I + ++ + + + R E + ER L K
Sbjct 508 SSARVKNASAYRTKFEATLPDIMESFNDLYRSI---------TGRITAEALK--ERVL-K 555
Query 121 MTKLWEAWGVYPPAFLRGIEATLCGRDITATILDPEAAAAAAAKCEAAALELADPQLDGE 180
+ ++W W ++ A++ G+ +T ++ + + C A P+++ +
Sbjct 556 VLQVWADWFLFSDAYIYGLRSTFLRSGVSGV-------TSFHSICGDA------PEIENK 602
Query 181 PLAENLRPL-----IAQFPLWLRPAAMEWLLLDRYQLDRLCQQRGLGVLPTR 227
A+N+ + A + A E + L +L+R C+ GL ++ R
Sbjct 603 SYADNMSDIGKINPDAALAIGKGAARQELMNLPIAELERRCRHNGLSLVGGR 654
> ath:AT5G10800 RNA recognition motif (RRM)-containing protein
Length=947
Score = 77.0 bits (188), Expect = 5e-14, Method: Composition-based stats.
Identities = 63/228 (27%), Positives = 108/228 (47%), Gaps = 22/228 (9%)
Query 1 LLRNITRDRASICAAMSFCMSHGSSSAEVCSCLTESLTLPDTALTMRLARMYLLSDLLAN 60
+LR +T +R+ I AM F + + ++ EV LTESLTL +T++ ++AR+ L+SD++ N
Sbjct 455 MLRALTLERSQIREAMGFALDNAEAAGEVVEVLTESLTLKETSIPTKVARLMLVSDIIHN 514
Query 61 SSAQTAHAWTYRHHLEKHLPFIADRWSKVLEDLRLGVGVGGASPRTQQEQWQGVERALYK 120
SSA+ +A YR E LP I + ++ + + R E + ER L K
Sbjct 515 SSARVKNASAYRTKFEATLPDIMESFNDLYHSVH---------GRITAEALR--ERVL-K 562
Query 121 MTKLWEAWGVYPPAFLRGIEAT-LCGRDITATILDPEAAAAAAAKCEAAALELADPQLDG 179
+ ++W W ++ A++ G+ AT L R+ T A + + + D
Sbjct 563 VLQVWADWFLFSDAYINGLRATFLRSRNFGVTSFHSICGDAPDIEKKGLIGNMNDADKIN 622
Query 180 EPLAENLRPLIAQFPLWLRPAAMEWLLLDRYQLDRLCQQRGLGVLPTR 227
+ A + A+ L RP + +L+R C+ GL +L R
Sbjct 623 QDAALAMGEGAARQELMNRPIS---------ELERRCRHNGLSLLGGR 661
> cel:C07H6.4 hypothetical protein; K12842 U2-associated protein
SR140
Length=933
Score = 65.9 bits (159), Expect = 1e-10, Method: Composition-based stats.
Identities = 39/135 (28%), Positives = 71/135 (52%), Gaps = 17/135 (12%)
Query 1 LLRNITRDRASICAAMSFCMSHGSSSAEVCSCLTESLTLPDTALTMRLARMYLLSDLLAN 60
L+R +T ++ SI A M +C+ + +AE+C C+ +SLT+ D L +++R+YL++D+L+N
Sbjct 533 LIRELTPEKTSIAATMVWCIQNAKYAAEICECVYDSLTIEDIPLYKKISRLYLINDILSN 592
Query 61 SSAQTAH-AWTYRHHLEKHLPFIADRWSKVLEDLRLGVGVG--GASPRTQQEQWQGVERA 117
+ + YR H E +LE + + VG R +QEQ++
Sbjct 593 CVQRNVRDVFLYRSHFE-----------ALLEKIFVAVGKAYRAIPSRIKQEQFKQRVMC 641
Query 118 LYKMTKLWEAWGVYP 132
+++ +E VYP
Sbjct 642 VFRN---FEEMAVYP 653
> tgo:TGME49_040710 RRM domain-containing protein
Length=859
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 34/83 (40%), Positives = 48/83 (57%), Gaps = 4/83 (4%)
Query 137 RGIEATLCGRDITATILDPEAAAAAAAKCEAAALELADPQLDGEPLAENLRPLIAQFPLW 196
+G+EA+L D A + EA A EAA EL D +DG+P+A L+A++PL
Sbjct 524 KGLEASLFSADFPAQLA--EAPEEAGPPAEAA--ELVDDDVDGQPIAAAEACLLAKYPLQ 579
Query 197 LRPAAMEWLLLDRYQLDRLCQQR 219
LR +W LDR +L++LC QR
Sbjct 580 LREQIHQWCRLDRSELEKLCLQR 602
> cpv:cgd8_1480 RPR domain containing protein, present in proteins
involved in mRNA splicing
Length=324
Score = 51.2 bits (121), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 40/212 (18%), Positives = 87/212 (41%), Gaps = 23/212 (10%)
Query 1 LLRNITRDRASICAAMSFCMSHGSS-SAEVCSCLTESLTLPDTALTMRLARMYLLSDLLA 59
LL ++T R I M+F + + S ++ L + P++ ++++ +Y +SD+L
Sbjct 91 LLDSLTTKRMKINEIMTFVVDYSDQHSVQMIELLIQRF--PNSTFEVKVSILYCISDILY 148
Query 60 NSSAQTAHAWTYRHHLEKHLPFIADRWSKVLEDLRLGVGVGGASPRTQQEQWQGVERALY 119
NS + AW R+ + P++ + S R + +
Sbjct 149 NSHSSKIGAWKLRNCIMNIFPYLVSH-------------ISFQSNRGNSCYIDLINKT-K 194
Query 120 KMTKLWEAWGVYPPAFLRGIEATLCGRDITATILDPEAAAAAAAKCEAAALELADPQLDG 179
+ ++W W ++P ++ G+ ++LC +LD E++D L
Sbjct 195 SIIEIWIDWKIFPSEYIEGLSSSLC----FDKMLDEMKNDKKTKLISNINDEISDGILLD 250
Query 180 EPLAENLR--PLIAQFPLWLRPAAMEWLLLDR 209
+ + L P+ ++ P+W M LL+++
Sbjct 251 KDIVSILSIWPIKSRLPVWKVWREMNNLLINK 282
> mmu:237504 Rassf9, AW322107, MGC41694, Pamci; Ras association
(RalGDS/AF-6) domain family (N-terminal) member 9
Length=435
Score = 32.7 bits (73), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 18/40 (45%), Positives = 23/40 (57%), Gaps = 3/40 (7%)
Query 99 VGGASPRTQQEQWQGVERALYKMT---KLWEAWGVYPPAF 135
+G AS E+W+G ERAL +T KLW+AWG P
Sbjct 71 LGKASDYCIVEKWRGSERALPPLTRILKLWKAWGDEQPNM 110
> dre:100150148 ras association domain-containing protein 9-like
Length=435
Score = 31.2 bits (69), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 22/34 (64%), Gaps = 4/34 (11%)
Query 109 EQWQGVERALYKMT---KLWEAWGVYPPAFLRGI 139
E+W+G ERAL +T +LW AWG P F++ +
Sbjct 81 ERWKGFERALPPLTRILRLWNAWGDEKP-FIKFV 113
> xla:100158456 rassf9, P-CIP1; Ras association (RalGDS/AF-6)
domain family (N-terminal) member 9
Length=420
Score = 30.4 bits (67), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 13/24 (54%), Positives = 17/24 (70%), Gaps = 3/24 (12%)
Query 109 EQWQGVERAL---YKMTKLWEAWG 129
E+W+G ER L K+ KLW+AWG
Sbjct 72 EKWRGFERVLPPPTKILKLWKAWG 95
Lambda K H
0.321 0.132 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9148686404
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40