bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0268_orf1
Length=117
Score E
Sequences producing significant alignments: (Bits) Value
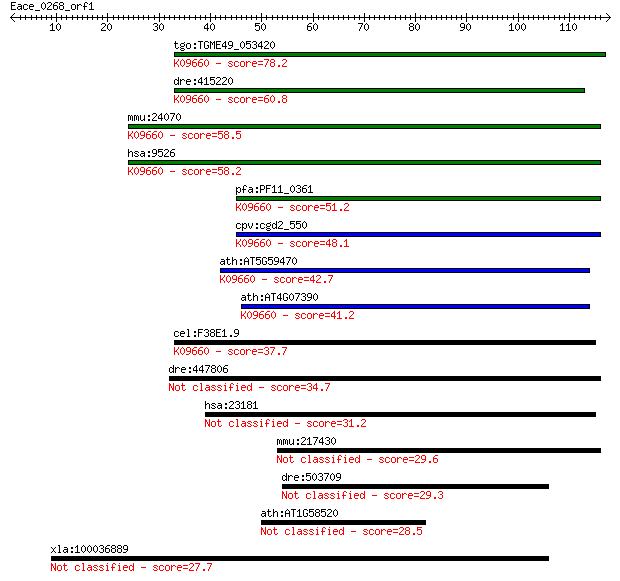
tgo:TGME49_053420 hypothetical protein ; K09660 mannose-P-doli... 78.2 5e-15
dre:415220 mpdu1b, fb18d03, wu:fb18d03, zgc:86765; mannose-P-d... 60.8 1e-09
mmu:24070 Mpdu1, LEC35, SL15, Supl15h; mannose-P-dolichol util... 58.5 5e-09
hsa:9526 MPDU1, CDGIF, FLJ14836, HBEBP2BPA, Lec35, My008, PP39... 58.2 6e-09
pfa:PF11_0361 conserved Plasmodium protein; K09660 mannose-P-d... 51.2 9e-07
cpv:cgd2_550 hypothetical protein ; K09660 mannose-P-dolichol ... 48.1 7e-06
ath:AT5G59470 PQ-loop repeat family protein / transmembrane fa... 42.7 3e-04
ath:AT4G07390 PQ-loop repeat family protein / transmembrane fa... 41.2 0.001
cel:F38E1.9 hypothetical protein; K09660 mannose-P-dolichol ut... 37.7 0.010
dre:447806 mpdu1a, mpdu1, zgc:92112; mannose-P-dolichol utiliz... 34.7 0.067
hsa:23181 DIP2A, C21orf106, DIP2; DIP2 disco-interacting prote... 31.2 0.74
mmu:217430 Pqlc3, C78076, E030024M05Rik; PQ loop repeat contai... 29.6 2.5
dre:503709 zgc:113491 29.3 2.9
ath:AT1G58520 RXW8; hydrolase, acting on ester bonds / lipase 28.5 5.3
xla:100036889 pqlc2; PQ loop repeat containing 2 27.7
> tgo:TGME49_053420 hypothetical protein ; K09660 mannose-P-dolichol
utilization defect 1
Length=286
Score = 78.2 bits (191), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 35/84 (41%), Positives = 54/84 (64%), Gaps = 0/84 (0%)
Query 33 CIKESAGLTLNVLILAGALIVKTPQLLNVFIAKSASGLSYWAILTEVFTAELVVGYNLLS 92
C K+ L++ IL G +VK PQL+ + A+S +GL+ ++ E +A + V YN+L
Sbjct 39 CAKQVVSACLSIGILVGGSLVKLPQLVKILRAQSVAGLAEMSVFVEAISASIFVAYNVLE 98
Query 93 KHPFSTWGEAVFIGAQNLVLLLLY 116
+HPF+TWGE +F+ QNL LLL+
Sbjct 99 RHPFTTWGEMLFVSLQNLCTLLLF 122
> dre:415220 mpdu1b, fb18d03, wu:fb18d03, zgc:86765; mannose-P-dolichol
utilization defect 1b; K09660 mannose-P-dolichol utilization
defect 1
Length=255
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 31/80 (38%), Positives = 51/80 (63%), Gaps = 0/80 (0%)
Query 33 CIKESAGLTLNVLILAGALIVKTPQLLNVFIAKSASGLSYWAILTEVFTAELVVGYNLLS 92
C+K L + I+ G+++VK PQ+L + AKSA GLS+ ++L E+F + Y+L +
Sbjct 44 CLKIVISKGLGIGIILGSVLVKLPQILKLLGAKSAEGLSFNSVLLELFAITGTMAYSLAN 103
Query 93 KHPFSTWGEAVFIGAQNLVL 112
PFS+WGEA+F+ Q + +
Sbjct 104 SFPFSSWGEALFLMFQTVTI 123
> mmu:24070 Mpdu1, LEC35, SL15, Supl15h; mannose-P-dolichol utilization
defect 1; K09660 mannose-P-dolichol utilization defect
1
Length=247
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 33/93 (35%), Positives = 55/93 (59%), Gaps = 1/93 (1%)
Query 24 YDRWEL-SSACIKESAGLTLNVLILAGALIVKTPQLLNVFIAKSASGLSYWAILTEVFTA 82
+ +W+L C+K L + I+AG+L+VK PQ+ + AKSA GLS +++ E+
Sbjct 27 FVQWDLLHVPCLKILLSKGLGLGIVAGSLLVKLPQVFKLLGAKSAEGLSLQSVMLELVAL 86
Query 83 ELVVGYNLLSKHPFSTWGEAVFIGAQNLVLLLL 115
V Y++ + PFS+WGEA+F+ Q + + L
Sbjct 87 TGTVVYSITNNFPFSSWGEALFLTLQTVAICFL 119
> hsa:9526 MPDU1, CDGIF, FLJ14836, HBEBP2BPA, Lec35, My008, PP3958,
PQLC5, SL15; mannose-P-dolichol utilization defect 1;
K09660 mannose-P-dolichol utilization defect 1
Length=247
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 55/93 (59%), Gaps = 1/93 (1%)
Query 24 YDRWEL-SSACIKESAGLTLNVLILAGALIVKTPQLLNVFIAKSASGLSYWAILTEVFTA 82
+ +W+L C+K L + I+AG+L+VK PQ+ + AKSA GLS +++ E+
Sbjct 27 FVQWDLLHVPCLKILLSKGLGLGIVAGSLLVKLPQVFKILGAKSAEGLSLQSVMLELVAL 86
Query 83 ELVVGYNLLSKHPFSTWGEAVFIGAQNLVLLLL 115
+ Y++ + PFS+WGEA+F+ Q + + L
Sbjct 87 TGTMVYSITNNFPFSSWGEALFLMLQTITICFL 119
> pfa:PF11_0361 conserved Plasmodium protein; K09660 mannose-P-dolichol
utilization defect 1
Length=233
Score = 51.2 bits (121), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 41/71 (57%), Gaps = 0/71 (0%)
Query 45 LILAGALIVKTPQLLNVFIAKSASGLSYWAILTEVFTAELVVGYNLLSKHPFSTWGEAVF 104
I+AGA +K PQL + K+A GLS+ +I E+F A ++ +++ K F + + +
Sbjct 30 FIIAGACFIKIPQLRKIITKKTAVGLSFMSIYLEIFVATSLIVFSIYEKINFILYVDVIL 89
Query 105 IGAQNLVLLLL 115
I QNL+L+
Sbjct 90 INVQNLILVFF 100
> cpv:cgd2_550 hypothetical protein ; K09660 mannose-P-dolichol
utilization defect 1
Length=244
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 43/71 (60%), Gaps = 0/71 (0%)
Query 45 LILAGALIVKTPQLLNVFIAKSASGLSYWAILTEVFTAELVVGYNLLSKHPFSTWGEAVF 104
LI+AG+ IVK PQ++ + ++S G+S ++I E+ ++ + N P+ W ++ F
Sbjct 30 LIIAGSCIVKIPQIIKILNSRSTQGISSFSIYVEILSSCIYSFSNWRFNVPWLLWADSAF 89
Query 105 IGAQNLVLLLL 115
IG QN +L+L
Sbjct 90 IGIQNAFILIL 100
> ath:AT5G59470 PQ-loop repeat family protein / transmembrane
family protein; K09660 mannose-P-dolichol utilization defect
1
Length=239
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 38/72 (52%), Gaps = 0/72 (0%)
Query 42 LNVLILAGALIVKTPQLLNVFIAKSASGLSYWAILTEVFTAELVVGYNLLSKHPFSTWGE 101
L ++A ++ VK PQ++ + KS GLS A EV + + Y L PFS +GE
Sbjct 34 LGYFLVAASMTVKLPQIMKIVDNKSVKGLSVVAFELEVIGYTISLAYCLNKDLPFSAFGE 93
Query 102 AVFIGAQNLVLL 113
F+ Q L+L+
Sbjct 94 LAFLLIQALILV 105
> ath:AT4G07390 PQ-loop repeat family protein / transmembrane
family protein; K09660 mannose-P-dolichol utilization defect
1
Length=235
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/68 (35%), Positives = 37/68 (54%), Gaps = 0/68 (0%)
Query 46 ILAGALIVKTPQLLNVFIAKSASGLSYWAILTEVFTAELVVGYNLLSKHPFSTWGEAVFI 105
++A ++ VK PQ++ + KS GLS A EV + + Y L PFS +GE F+
Sbjct 38 LVAASITVKLPQIMKIVQHKSVRGLSVVAFELEVVGYTISLAYCLHKGLPFSAFGEMAFL 97
Query 106 GAQNLVLL 113
Q L+L+
Sbjct 98 LIQALILV 105
> cel:F38E1.9 hypothetical protein; K09660 mannose-P-dolichol
utilization defect 1
Length=238
Score = 37.7 bits (86), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 42/82 (51%), Gaps = 0/82 (0%)
Query 33 CIKESAGLTLNVLILAGALIVKTPQLLNVFIAKSASGLSYWAILTEVFTAELVVGYNLLS 92
C K L I G++++ PQ+L + A+SA G+S + L + A Y+ S
Sbjct 28 CPKAVLSRGLGFAITLGSILLFVPQILKIQAARSAQGISAASQLLALVGAIGTASYSYRS 87
Query 93 KHPFSTWGEAVFIGAQNLVLLL 114
FS WG++ F+ Q ++++L
Sbjct 88 GFVFSGWGDSFFVAVQLVIIIL 109
> dre:447806 mpdu1a, mpdu1, zgc:92112; mannose-P-dolichol utilization
defect 1a
Length=258
Score = 34.7 bits (78), Expect = 0.067, Method: Compositional matrix adjust.
Identities = 21/84 (25%), Positives = 36/84 (42%), Gaps = 0/84 (0%)
Query 32 ACIKESAGLTLNVLILAGALIVKTPQLLNVFIAKSASGLSYWAILTEVFTAELVVGYNLL 91
C+K T+ + IL G +I PQ+ + S+ GL ++ ++ +
Sbjct 40 PCLKIVLSKTMGIFILMGIVIAPLPQICKILWCGSSYGLCLTSVFLDLMAISTHAAFCYT 99
Query 92 SKHPFSTWGEAVFIGAQNLVLLLL 115
P WGE++F Q +L LL
Sbjct 100 QNFPIGAWGESLFAVIQIALLALL 123
> hsa:23181 DIP2A, C21orf106, DIP2; DIP2 disco-interacting protein
2 homolog A (Drosophila)
Length=1110
Score = 31.2 bits (69), Expect = 0.74, Method: Composition-based stats.
Identities = 24/81 (29%), Positives = 41/81 (50%), Gaps = 5/81 (6%)
Query 39 GLTLNVLILAGA-LIVKTPQLLNVFIAKSASGLSYWAIL---TEVFTA-ELVVGYNLLSK 93
G + V+ L G + KT ++ + ++ SA+G +Y+ +L VF A + G +
Sbjct 661 GANVCVVKLEGTPYLCKTDEVGEICVSSSATGTAYYGLLGITKNVFEAVPVTTGGAPIFD 720
Query 94 HPFSTWGEAVFIGAQNLVLLL 114
PF+ G FIG NLV ++
Sbjct 721 RPFTRTGLLGFIGPDNLVFIV 741
> mmu:217430 Pqlc3, C78076, E030024M05Rik; PQ loop repeat containing
Length=170
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 34/63 (53%), Gaps = 0/63 (0%)
Query 53 VKTPQLLNVFIAKSASGLSYWAILTEVFTAELVVGYNLLSKHPFSTWGEAVFIGAQNLVL 112
+K PQ+ A+SA G+S ++L E+ + + Y +P T+ E + AQ++VL
Sbjct 20 LKLPQIYAQLAARSARGISLPSLLLELAGFLVFLRYQHYYGNPLLTYLEYPILIAQDIVL 79
Query 113 LLL 115
LL
Sbjct 80 LLF 82
> dre:503709 zgc:113491
Length=302
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 26/52 (50%), Gaps = 1/52 (1%)
Query 54 KTPQLLNVFIAKSASGLSYWAILTEVFTAELVVGYNLLSKHPFSTWGEAVFI 105
+ PQ+ F KS GLSY+ + V G ++L K+P GEA ++
Sbjct 205 RLPQIYTNFQRKSTEGLSYF-LFALVILGNTTYGVSVLLKNPDPGQGEASYM 255
> ath:AT1G58520 RXW8; hydrolase, acting on ester bonds / lipase
Length=1041
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 20/34 (58%), Gaps = 2/34 (5%)
Query 50 ALIVKTPQLLNVFIAKSASGLSYWAIL--TEVFT 81
A ++ Q+LNV+I K SG YW I T +F+
Sbjct 871 AYLIYKNQILNVYITKYESGGQYWPIFHNTTIFS 904
> xla:100036889 pqlc2; PQ loop repeat containing 2
Length=302
Score = 27.7 bits (60), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 25/103 (24%), Positives = 40/103 (38%), Gaps = 7/103 (6%)
Query 9 VLQSCGLASAGCAAAYDRWELSSA------CIKESAGLTLNVLILAGALIVKTPQLLNVF 62
+LQ G + A + L S +K G ++ LI + PQ+ F
Sbjct 152 LLQEAGSSPLNSADPFHSRHLLSTDGEEEYSVKNKIGFACGLMSTLSYLISRLPQIYTNF 211
Query 63 IAKSASGLSYWAILTEVFTAELVVGYNLLSKHPFSTWGEAVFI 105
KS GL+ L V + G ++L K+P S E ++
Sbjct 212 KRKSTEGLAPTLFLL-VIVGNVTYGASVLLKNPESDQSEGTYV 253
Lambda K H
0.324 0.137 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2032807080
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40