bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0171_orf1
Length=217
Score E
Sequences producing significant alignments: (Bits) Value
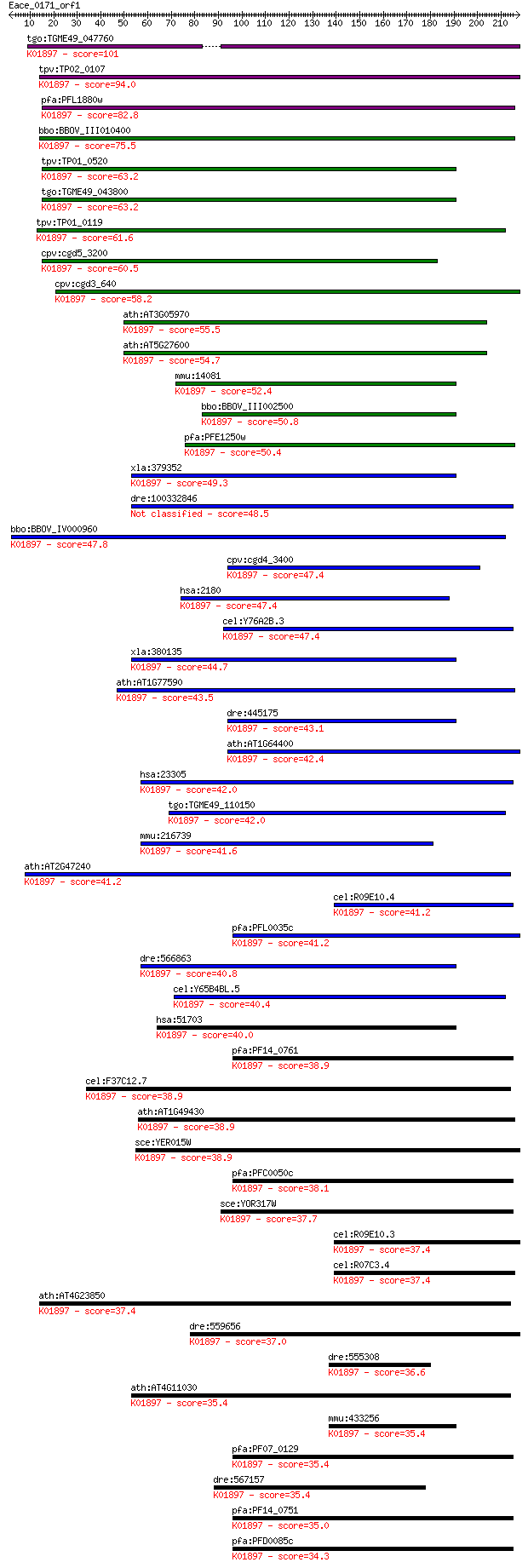
tgo:TGME49_047760 long chain acyl-CoA synthetase, putative (EC... 101 2e-21
tpv:TP02_0107 long-chain fatty acid CoA ligase; K01897 long-ch... 94.0 4e-19
pfa:PFL1880w ACS11; acyl-CoA synthetase, PfACS11 (EC:6.2.1.3);... 82.8 8e-16
bbo:BBOV_III010400 17.m07898; long-chain acyl-CoA synthetase (... 75.5 1e-13
tpv:TP01_0520 long-chain fatty acid CoA ligase; K01897 long-ch... 63.2 6e-10
tgo:TGME49_043800 long-chain fatty acid CoA ligase, putative (... 63.2 8e-10
tpv:TP01_0119 long-chain fatty acid CoA ligase; K01897 long-ch... 61.6 2e-09
cpv:cgd5_3200 acyl-CoA synthetase ; K01897 long-chain acyl-CoA... 60.5 4e-09
cpv:cgd3_640 acyl-CoA synthetase ; K01897 long-chain acyl-CoA ... 58.2 2e-08
ath:AT3G05970 LACS6; LACS6 (long-chain acyl-CoA synthetase 6);... 55.5 1e-07
ath:AT5G27600 LACS7; LACS7 (LONG-CHAIN ACYL-COA SYNTHETASE 7);... 54.7 3e-07
mmu:14081 Acsl1, Acas, Acas1, Acs, FACS, Facl2; acyl-CoA synth... 52.4 1e-06
bbo:BBOV_III002500 17.m07241; fatty acyl-CoA synthetase family... 50.8 3e-06
pfa:PFE1250w PfACS10; acyl-CoA synthetase, PfACS10 (EC:6.2.1.3... 50.4 4e-06
xla:379352 MGC53832; similar to fatty acid Coenzyme A ligase; ... 49.3 1e-05
dre:100332846 acyl-CoA synthetase long-chain family member 6-like 48.5 2e-05
bbo:BBOV_IV000960 21.m02785; fatty acyl-CoA synthetase 3 (EC:6... 47.8 3e-05
cpv:cgd4_3400 long chain fatty acyl CoA synthetase having a si... 47.4 4e-05
hsa:2180 ACSL1, ACS1, FACL1, FACL2, LACS, LACS1, LACS2; acyl-C... 47.4 4e-05
cel:Y76A2B.3 acs-14; fatty Acid CoA Synthetase family member (... 47.4 4e-05
xla:380135 acsl1, MGC132122, MGC52902, facl2; acyl-CoA synthet... 44.7 2e-04
ath:AT1G77590 LACS9; LACS9 (LONG CHAIN ACYL-COA SYNTHETASE 9);... 43.5 6e-04
dre:445175 zgc:101071 (EC:6.2.1.3); K01897 long-chain acyl-CoA... 43.1 8e-04
ath:AT1G64400 long-chain-fatty-acid--CoA ligase, putative / lo... 42.4 0.001
hsa:23305 ACSL6, ACS2, FACL6, FLJ16173, KIAA0837, LACS_6, LACS... 42.0 0.002
tgo:TGME49_110150 long-chain-fatty-acid-CoA ligase, putative (... 42.0 0.002
mmu:216739 Acsl6, A330035H04Rik, AW050338, Facl6, LACS, Lacsl,... 41.6 0.002
ath:AT2G47240 long-chain-fatty-acid--CoA ligase family protein... 41.2 0.003
cel:R09E10.4 hypothetical protein; K01897 long-chain acyl-CoA ... 41.2 0.003
pfa:PFL0035c ACS7; acyl-CoA synthetase, PfACS7; K01897 long-ch... 41.2 0.003
dre:566863 fa04h02, wu:fc05h07; wu:fa04h02; K01897 long-chain ... 40.8 0.004
cel:Y65B4BL.5 acs-13; fatty Acid CoA Synthetase family member ... 40.4 0.005
hsa:51703 ACSL5, ACS2, ACS5, FACL5; acyl-CoA synthetase long-c... 40.0 0.006
pfa:PF14_0761 ACS1a; acyl-CoA synthetase; K01897 long-chain ac... 38.9 0.012
cel:F37C12.7 acs-16; fatty Acid CoA Synthetase family member (... 38.9 0.013
ath:AT1G49430 LACS2; LACS2 (LONG-CHAIN ACYL-COA SYNTHETASE 2);... 38.9 0.014
sce:YER015W FAA2, FAM1; Faa2p (EC:6.2.1.3); K01897 long-chain ... 38.9 0.016
pfa:PFC0050c PfACS2; acyl-CoA synthetase, PfACS2 (EC:6.2.1.3);... 38.1 0.023
sce:YOR317W FAA1; Faa1p (EC:6.2.1.3); K01897 long-chain acyl-C... 37.7 0.030
cel:R09E10.3 hypothetical protein; K01897 long-chain acyl-CoA ... 37.4 0.040
cel:R07C3.4 hypothetical protein; K01897 long-chain acyl-CoA s... 37.4 0.042
ath:AT4G23850 long-chain-fatty-acid--CoA ligase / long-chain a... 37.4 0.043
dre:559656 acsl3b, acsl3, fb34c03, im:7155129, si:dkey-20f20.5... 37.0 0.046
dre:555308 acsl1, MGC110081, zgc:110081; acyl-CoA synthetase l... 36.6 0.064
ath:AT4G11030 long-chain-fatty-acid--CoA ligase, putative / lo... 35.4 0.13
mmu:433256 Acsl5, 1700030F05Rik, ACS2, ACS5, Facl5; acyl-CoA s... 35.4 0.15
pfa:PF07_0129 PfACS5; acyl-coA synthetase, PfACS5 (EC:6.2.1.3)... 35.4 0.15
dre:567157 acsl3a, si:dkeyp-109h9.2; acyl-CoA synthetase long-... 35.4 0.17
pfa:PF14_0751 ACS1b; acyl-CoA synthetase, PfACS1b; K01897 long... 35.0 0.23
pfa:PFD0085c PfACS6; acyl-CoA synthetase, PfACS6 (EC:6.2.1.3);... 34.3 0.30
> tgo:TGME49_047760 long chain acyl-CoA synthetase, putative (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=793
Score = 101 bits (252), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 52/128 (40%), Positives = 80/128 (62%), Gaps = 3/128 (2%)
Query 91 LSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWR 150
++K+ +Y +TL+ V++L AL EV +P+ + ++ + R G+ +R+ WR
Sbjct 193 FGEYKWLSYQETLKQVQSLAWALSNEVDIPVCLFGDDA--EVQENYRFAGIWARSSAAWR 250
Query 151 LLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAALQL-KKEGFPLKA 209
+ D+A N IV+VPLYDTLG ++ +I+ LTK++VL EG+KL AL L +E PLKA
Sbjct 251 ITDYACNAAKIVSVPLYDTLGHEALLYIIGLTKLQVLFVEGAKLHPALNLVTEEKVPLKA 310
Query 210 VISFDEPT 217
VI FD T
Sbjct 311 VICFDSVT 318
Score = 39.7 bits (91), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 44/78 (56%), Gaps = 7/78 (8%)
Query 9 KTFKGKYALPIEGTETADS-SAVYRAVPDPSN--PTQTMLGEPAIPQ-FPHIKSTYPLLK 64
+ + G YA+P+EG + SAVYR V + P +T+ PA+ + FP I+S + L
Sbjct 22 RAWSGIYAVPVEGVPKKEGESAVYRCVESEQSGVPVKTL---PAVLEDFPEIRSPFDTLY 78
Query 65 EAARIFGRNLYVGERQPI 82
AA+ F + ++G R+ +
Sbjct 79 CAAQAFSNDPFLGVREKL 96
> tpv:TP02_0107 long-chain fatty acid CoA ligase; K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=697
Score = 94.0 bits (232), Expect = 4e-19, Method: Composition-based stats.
Identities = 66/204 (32%), Positives = 105/204 (51%), Gaps = 8/204 (3%)
Query 14 KYALPIEGTETADSSAVYRAVPDPSNPTQTMLGEPAIPQFPHIKSTYPLLKEAARIFGRN 73
KY+ + + +D S VY + D + +T + IP HI S++ +L E A+ FG N
Sbjct 27 KYSKFKQKSNRSDESDVYCPIDDDNLSPKT---KAIIPD--HINSSFDMLCETAKYFGNN 81
Query 74 LYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAP 133
++G R+ I + + ++ + TY D+++ + +G AL +E +P S D
Sbjct 82 NHLGVREKITLPDGKCEFGNYVFNTYNDSVKYAKIIGTALTKENLIPESTIDNCRF---V 138
Query 134 KPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSK 193
K R LG+ S W L D+A G V+VP+Y+TLG+ ++ I+K TKMEV + K
Sbjct 139 KKARFLGIWSMNCPYWLLTDYACCGYGFVSVPIYETLGDDALFKIIKTTKMEVACIDSKK 198
Query 194 LDAALQLKKEGFPLKAVISFDEPT 217
+ L E LK VI FD+ T
Sbjct 199 ISNLEHLFSEFKQLKKVIVFDQLT 222
> pfa:PFL1880w ACS11; acyl-CoA synthetase, PfACS11 (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=792
Score = 82.8 bits (203), Expect = 8e-16, Method: Composition-based stats.
Identities = 58/205 (28%), Positives = 97/205 (47%), Gaps = 14/205 (6%)
Query 15 YALPIEGTETADSSAVYR----AVPDPSNPTQTMLGEPAIPQFPHIKSTYPLLKEAARIF 70
Y I+GT +SS +YR + T LG PA S Y + + +
Sbjct 34 YFEEIKGTGKKNSSNIYRPYNYKINSEFEKTTGFLGIPA-------SSKYEVFCFFCKYY 86
Query 71 GRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLN 130
+GER+ ++ L + + TYA +E AL Q + + +++ G+
Sbjct 87 YNKDLLGERKKEIRGDNDVVLGGYTFKTYAQVKNEIEIFASALYQFEEIEFNIFNDNGIY 146
Query 131 PAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAE 190
++LG+ S+ R +W + D A + VTVP+YDT+G SV +I K T M+V E
Sbjct 147 ---NQFKILGIWSKNRSEWFITDLATSAINFVTVPIYDTIGLNSVIYIFKKTNMKVCCIE 203
Query 191 GSKLDAALQLKKEGFPLKAVISFDE 215
KL++ +++K+E L +I +DE
Sbjct 204 AEKLESLIKMKEELVDLSILIIYDE 228
> bbo:BBOV_III010400 17.m07898; long-chain acyl-CoA synthetase
(EC:6.2.1.3); K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=687
Score = 75.5 bits (184), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 60/204 (29%), Positives = 100/204 (49%), Gaps = 13/204 (6%)
Query 14 KYALPIEGTETADSSAVYRAVPDPSNPTQTMLGEPAIPQFPH-IKSTYPLLKEAARIFGR 72
+Y+ I D S VY A+ + IP FP ++++ +L+ A
Sbjct 26 QYSRVICQNPKKDESDVYCALNNKGECVD-------IPPFPERFQTSFDMLRHMAEQNPN 78
Query 73 NLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPA 132
++G R+ +S + TL ++K++T + T LG A E G+ + E L+ A
Sbjct 79 RDHLGVREKHVNSKGEITLGEYKWYTISYTATTAVILGSAFVSEPGL----FSETVLDDA 134
Query 133 P-KPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEG 191
K + LG+ + W + D+AA G VTVPLY+T+G+ ++ I K TKM+ L +
Sbjct 135 ILKKAKFLGIWATNCPLWLISDYAAIAFGFVTVPLYETMGDEAILTIFKETKMKTLCIDA 194
Query 192 SKLDAALQLKKEGFPLKAVISFDE 215
+KL L+LK +K +I FD+
Sbjct 195 AKLPTLLKLKDRLPEVKNLILFDK 218
> tpv:TP01_0520 long-chain fatty acid CoA ligase; K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=669
Score = 63.2 bits (152), Expect = 6e-10, Method: Composition-based stats.
Identities = 45/179 (25%), Positives = 83/179 (46%), Gaps = 20/179 (11%)
Query 15 YALPIEGTETADSSAVYRAVPDPSNPTQTMLGEPAIPQFPHIKSTYPLLKEAARIFGRNL 74
Y++ +EGTE + VYR +P + + E ++ IF R L
Sbjct 5 YSVALEGTEEEGYTPVYR---NPDYKDRLLSCEDYFDG---------KIQTGWDIFNRGL 52
Query 75 YVGERQPIKDS---NSQTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNP 131
+ +P S N+ T +FK+ TY + ++ G L + L + E +
Sbjct 53 TLSRNKPFLGSRVKNADGTFGEFKFMTYGEVEDQIKHFGSGL-----LNLDKFKEVFVKE 107
Query 132 APKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAE 190
+ +++LG+ S+ ++W + + N + VPLYDTLGE S+ +I+ +TK+ V+ +
Sbjct 108 ENQMVKMLGIYSQNTVEWLITEQVCNGYNLTLVPLYDTLGEESLLYIINVTKLNVIVCD 166
> tgo:TGME49_043800 long-chain fatty acid CoA ligase, putative
(EC:6.2.1.3); K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=748
Score = 63.2 bits (152), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 50/180 (27%), Positives = 80/180 (44%), Gaps = 18/180 (10%)
Query 15 YALPIEGTETADS--SAVYRAVPDPSNPTQTMLGEPAIPQFPHIKSTYPLLKEAARIFGR 72
Y +PIE A +++YR+V P P +S + L + I G
Sbjct 12 YNVPIEDPVPAGKKRTSIYRSVFSPDAIVDNFADRPC-------QSAWDLFQRGVAISGE 64
Query 73 NLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRA-LDQEVGVPLSHYDEEGLNP 131
+G R N+ TL +++ TY + ++ +G L P H++EE
Sbjct 65 KRCLGTRV----KNADGTLGPYQWKTYREVEQLALEVGSGILSLPDAAPKLHFEEETFQ- 119
Query 132 APKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKM-EVLAAE 190
+ +R L S+ R +W + + A N GI VPLYDTLG S I+ T++ V+A+E
Sbjct 120 --QDMRFLAFYSKNREEWAICEQACNAYGITIVPLYDTLGPESTAFILAQTRLRSVVASE 177
> tpv:TP01_0119 long-chain fatty acid CoA ligase; K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=691
Score = 61.6 bits (148), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 56/202 (27%), Positives = 91/202 (45%), Gaps = 18/202 (8%)
Query 13 GKYALPIEGTETADSSAVYRAVPDPSNPTQTMLGEPAIPQFPHIKSTYPLLKEAARIFGR 72
G Y++PI GTE S VYR P + + +LG F K+++ L + R
Sbjct 10 GFYSVPIPGTEEEGFSPVYR---HPKH--EKILGAS---DFGDFKTSWDLFQAGLRRNPD 61
Query 73 NLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPA 132
VG+R+ + D L DF++ TY++ + +A+G +L V +
Sbjct 62 AECVGKRKRLPDGK----LGDFEFNTYSEFFKTTKAVGSSLVHHNLVKEQRISQSSFQGT 117
Query 133 PKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAA--- 189
K ++GL + +W LL+ A G VP+Y TLG S+ ++ T +E+L
Sbjct 118 CK---LVGLFLPSCEEWLLLEQACYGYGYTLVPIYTTLGTESILFVLTNTGLELLFCTEE 174
Query 190 EGSKLDAALQLKKEGFPLKAVI 211
KL L L K PL+ ++
Sbjct 175 NAEKLFEVLSLSKTKLPLRNLV 196
> cpv:cgd5_3200 acyl-CoA synthetase ; K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=683
Score = 60.5 bits (145), Expect = 4e-09, Method: Composition-based stats.
Identities = 41/168 (24%), Positives = 76/168 (45%), Gaps = 16/168 (9%)
Query 15 YALPIEGTETADSSAVYRAVPDPSNPTQTMLGEPAIPQFPHIKSTYPLLKEAARIFGRNL 74
Y+ PIEGT ++S+ + R +T+ E +++ + +L
Sbjct 17 YSCPIEGTGDSNSTEIRRC-------KETINSELTSNFHEKLENCWGILLHGKEASNNGD 69
Query 75 YVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPK 134
Y+G R P+K+ LSD+K+ Y+ +R + +G L + + E
Sbjct 70 YMGVRVPLKNGE----LSDYKFVKYSYVIRKAKEVGSGL-----LHIGAVQERSFEDCNI 120
Query 135 PLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLT 182
L+ + L ++ ++W + + A N GI T+PLYD LG + +I+ T
Sbjct 121 KLKCVALFAKNSLNWSITEQACNAYGISTIPLYDVLGNSGLTYILNST 168
> cpv:cgd3_640 acyl-CoA synthetase ; K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=685
Score = 58.2 bits (139), Expect = 2e-08, Method: Composition-based stats.
Identities = 50/205 (24%), Positives = 93/205 (45%), Gaps = 22/205 (10%)
Query 21 GTETADSSAVYRAVPDPSNPTQTMLGEPAIPQFPHIKSTYPLLKEAARIFGRNLYVGERQ 80
G +SS +YR NP+ + GE + Q + + + L + + +G R+
Sbjct 18 GQVEPESSPIYR------NPSYSK-GELSNLQGLNANNLWELFVNSVNKYKNKKCLGTRK 70
Query 81 PIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPLRVLG 140
+D T ++ + +Y + +G + + P+ Y++ N K + ++G
Sbjct 71 LNRDG----TYGEYVFKSYEELKEEALNIGINILKMDLCPIRKYED---NEYQKEISMMG 123
Query 141 LRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAALQL 200
+ SR R +W L + A N GI PLYDTLGE ++K I+ T+++ L L+ + +
Sbjct 124 ILSRNREEWYLTEHACNAFGICLAPLYDTLGEDNLKFILVQTQLKSLCISNDSLEKIISI 183
Query 201 K--------KEGFPLKAVISFDEPT 217
++ +K +I FD PT
Sbjct 184 IERSIIDSTRDSILIKNLICFDHPT 208
> ath:AT3G05970 LACS6; LACS6 (long-chain acyl-CoA synthetase 6);
long-chain-fatty-acid-CoA ligase (EC:6.2.1.3); K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=701
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 40/154 (25%), Positives = 64/154 (41%), Gaps = 20/154 (12%)
Query 50 IPQFPHIKSTYPLLKEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEAL 109
P P I + + + A F Y+G R + T+ D+K+ TY + AL
Sbjct 77 FPDHPDIATLHDNFEHAVHDFRDYKYLGTRVRV-----DGTVGDYKWMTYGEAGTARTAL 131
Query 110 GRALDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDT 169
G L G+P+ +G+ R +W ++D A + V+VPLYDT
Sbjct 132 GSGLVHH-GIPMG--------------SSVGIYFINRPEWLIVDHACSSYSYVSVPLYDT 176
Query 170 LGEPSVKHIVKLTKMEVLAAEGSKLDAALQLKKE 203
LG +VK IV ++ + L++ L E
Sbjct 177 LGPDAVKFIVNHATVQAIFCVAETLNSLLSCLSE 210
> ath:AT5G27600 LACS7; LACS7 (LONG-CHAIN ACYL-COA SYNTHETASE 7);
long-chain-fatty-acid-CoA ligase/ protein binding (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=700
Score = 54.7 bits (130), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 39/154 (25%), Positives = 62/154 (40%), Gaps = 20/154 (12%)
Query 50 IPQFPHIKSTYPLLKEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEAL 109
P P I + + A + N Y+G R S T+ ++ + TY + +A+
Sbjct 77 FPDHPEIGTLHDNFVHAVETYAENKYLGTR-----VRSDGTIGEYSWMTYGEAASERQAI 131
Query 110 GRALDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDT 169
G L L H +G +GL R +W ++D A V+VPLYDT
Sbjct 132 GSGL-------LFHGVNQG--------DCVGLYFINRPEWLVVDHACAAYSFVSVPLYDT 176
Query 170 LGEPSVKHIVKLTKMEVLAAEGSKLDAALQLKKE 203
LG +VK +V ++ + L+ L E
Sbjct 177 LGPDAVKFVVNHANLQAIFCVPQTLNILLSFLAE 210
> mmu:14081 Acsl1, Acas, Acas1, Acs, FACS, Facl2; acyl-CoA synthetase
long-chain family member 1 (EC:6.2.1.3); K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=699
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 32/125 (25%), Positives = 61/125 (48%), Gaps = 19/125 (15%)
Query 72 RNLYVGERQPIKDSNSQTTLSD------FKYFTYADTLRMVEALGRALDQEVGVPLSHYD 125
R +Y G ++ I+ SN+ L +++ +Y + + E +G L Q+
Sbjct 91 RTMYDGFQRGIQVSNNGPCLGSRKPNQPYEWISYKEVAELAECIGSGLIQK--------- 141
Query 126 EEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKME 185
G P + + +GL S+ R +W +++ +V VPLYDTLG ++ +IV ++
Sbjct 142 --GFKPCSE--QFIGLFSQNRPEWVIVEQGCFSYSMVVVPLYDTLGADAITYIVNKAELS 197
Query 186 VLAAE 190
V+ A+
Sbjct 198 VIFAD 202
> bbo:BBOV_III002500 17.m07241; fatty acyl-CoA synthetase family
protein; K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=663
Score = 50.8 bits (120), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 25/108 (23%), Positives = 54/108 (50%), Gaps = 4/108 (3%)
Query 83 KDSNSQTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPLRVLGLR 142
++ + +L ++ + +Y + +V+ G L GV E L P P ++G+
Sbjct 61 REKKADGSLGEYVFKSYKEVESLVQRFGSGLRSLKGVN----KVEVLAPEPVEATMIGIY 116
Query 143 SRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAE 190
+ ++W + + N G VP+YDT+GE S+ HI++ + + ++ +
Sbjct 117 ASNCVEWLICEQTCNGYGYTIVPIYDTIGEESIIHILENSDINIVVCD 164
> pfa:PFE1250w PfACS10; acyl-CoA synthetase, PfACS10 (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=673
Score = 50.4 bits (119), Expect = 4e-06, Method: Composition-based stats.
Identities = 38/143 (26%), Positives = 69/143 (48%), Gaps = 12/143 (8%)
Query 76 VGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKP 135
+G R+ ++D+ +++ + + ++ +G L PL D++ + A
Sbjct 64 MGVREKLEDNKRGA----YQWKNFGEVKELIMKVGSGLLNMNACPLIMCDDQRIPKA--- 116
Query 136 LRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGS--- 192
R LGL +W + D N I+TVPLYD+LG S + I+ T+ME + + +
Sbjct 117 -RFLGLYMPNCPEWNICDLGCNAYNIITVPLYDSLGPQSSRFILDQTQMETIVCDKTCAR 175
Query 193 KLDAALQLKKEGFPLKAVISFDE 215
L +L+ +E + LK +I DE
Sbjct 176 NLFKSLETCEEIY-LKTLILVDE 197
> xla:379352 MGC53832; similar to fatty acid Coenzyme A ligase;
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=698
Score = 49.3 bits (116), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 28/138 (20%), Positives = 61/138 (44%), Gaps = 22/138 (15%)
Query 53 FPHIKSTYPLLKEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRA 112
+ +K+ Y + + R+ +G R+P + +++ +Y + E +G A
Sbjct 86 YDDVKTIYDIFQRGVRVSNNGPCLGHRKPNQP---------YEWISYKEVAERAEFVGSA 136
Query 113 LDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGE 172
L G P+P + +G+ ++ R +W +++ +V VPLYDTLG
Sbjct 137 LLHR-----------GFKPSPD--QFIGIFAQNRPEWTIVELGCYTYSMVAVPLYDTLGA 183
Query 173 PSVKHIVKLTKMEVLAAE 190
++ +I+ + V+ +
Sbjct 184 EAITYIINRADLSVVFCD 201
> dre:100332846 acyl-CoA synthetase long-chain family member 6-like
Length=1391
Score = 48.5 bits (114), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 36/165 (21%), Positives = 74/165 (44%), Gaps = 25/165 (15%)
Query 53 FPHIKSTYPLLKEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRA 112
+ K+ Y + + RI G +G R+P + +++ +Y + + LG
Sbjct 800 YEDTKTAYDMFQRGLRIAGDGPCLGFRKPREP---------YQWISYTEVAERAQVLGSG 850
Query 113 LDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGE 172
L + P P + +G+ ++ R +W + + A + VPLYDTLG
Sbjct 851 LLAK-------------GCKPNPQQYVGIYAQNRPEWVIAELACYTFSMALVPLYDTLGI 897
Query 173 PSVKHIVKLTKME-VLAAEGSKLDAALQLKKEGF--PLKAVISFD 214
++ HI+ L ++ V+ + K ++ L+ K++G L ++ F+
Sbjct 898 EAMVHILNLAEISMVICDKEDKAESLLRNKEKGVTPSLSCLVLFN 942
> bbo:BBOV_IV000960 21.m02785; fatty acyl-CoA synthetase 3 (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=681
Score = 47.8 bits (112), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 50/216 (23%), Positives = 86/216 (39%), Gaps = 23/216 (10%)
Query 2 LRQLLQVKTFK---GKYALPIEGTETADSSAVYRAVPDPSNPTQTMLGEPAIPQFPHIKS 58
+ LLQ + G YA+P+ T S +YR + +T L + + +
Sbjct 1 MENLLQYSNYGSVPGFYAVPVPDTAQTGYSPIYRNA--AVDTIKTAL------DYGNFHN 52
Query 59 TYPLLKEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQEVG 118
+ + + + +G+R N T + + TY +G AL Q +
Sbjct 53 MWDVFQHGLKKNPDAPCIGKRA----VNPDGTRGAYSFMTYKQVETFALLIGSALSQLLI 108
Query 119 VPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHI 178
P D P K R++GL ++W +L+ A N VP+Y+TLG S+ I
Sbjct 109 EPQISED-----PTVKDARMVGLFVPNCLEWLILEQACNAYAYTLVPIYNTLGHESIHTI 163
Query 179 VKLTKMEVLAAEGSKLD---AALQLKKEGFPLKAVI 211
+ +++ VL + L+ EG LK +I
Sbjct 164 LLNSRISVLLCTPDTVKIMFTVLEQGTEGVELKTII 199
> cpv:cgd4_3400 long chain fatty acyl CoA synthetase having a
signal peptide ; K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=766
Score = 47.4 bits (111), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 26/108 (24%), Positives = 47/108 (43%), Gaps = 5/108 (4%)
Query 94 FKYFTYADTLRMVEALGRAL-DQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLL 152
++F Y+ L++ G L + P + + KP+R + + S+ R++W L
Sbjct 128 MEWFRYSSVLKLAAEFGSGLLSLKDSAPFVDHSPD----IKKPIRTVSILSKNRLEWSLT 183
Query 153 DFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAALQL 200
+ A + G+V P+YD LG V H + L + L L +
Sbjct 184 EIACSTYGMVLSPMYDALGPDGVAHSINLVGSSTVVVSMEALSTILNV 231
> hsa:2180 ACSL1, ACS1, FACL1, FACL2, LACS, LACS1, LACS2; acyl-CoA
synthetase long-chain family member 1 (EC:6.2.1.3); K01897
long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=698
Score = 47.4 bits (111), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 30/120 (25%), Positives = 58/120 (48%), Gaps = 19/120 (15%)
Query 74 LYVGERQPIKDSNSQTTLSD------FKYFTYADTLRMVEALGRALDQEVGVPLSHYDEE 127
LY G ++ I+ SN+ L +++ +Y + E +G AL Q+
Sbjct 92 LYEGFQRGIQVSNNGPCLGSRKPDQPYEWLSYKQVAELSECIGSALIQK----------- 140
Query 128 GLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVL 187
G AP + +G+ ++ R +W +++ +V VPLYDTLG ++ +IV ++ ++
Sbjct 141 GFKTAPD--QFIGIFAQNRPEWVIIEQGCFAYSMVIVPLYDTLGNEAITYIVNKAELSLV 198
> cel:Y76A2B.3 acs-14; fatty Acid CoA Synthetase family member
(acs-14); K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=687
Score = 47.4 bits (111), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 32/124 (25%), Positives = 53/124 (42%), Gaps = 13/124 (10%)
Query 92 SDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRL 151
+D+++ TY D + L L E G+ P +G+ +R W +
Sbjct 100 ADYEFLTYDDVHEQAKNLSMTLVHEFGL------------TPANTTNIGIYARNSPQWLV 147
Query 152 LDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAALQLKKEGFP-LKAV 210
A + +V VPLYDTLG + I+ ++ V+ + K +L +E P LK +
Sbjct 148 SAVACVEQSMVVVPLYDTLGAEAATFIISQAEISVVIVDSFKKAESLIKNRENMPTLKNI 207
Query 211 ISFD 214
I D
Sbjct 208 IVID 211
> xla:380135 acsl1, MGC132122, MGC52902, facl2; acyl-CoA synthetase
long-chain family member 1 protein (EC:6.2.1.3); K01897
long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=698
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 27/138 (19%), Positives = 60/138 (43%), Gaps = 22/138 (15%)
Query 53 FPHIKSTYPLLKEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRA 112
+ +K+ Y + + R+ +G R+P + +++ +Y + E +G A
Sbjct 86 YEDVKTIYDIFQRGMRVSNNGPCLGHRKPNQP---------YEWISYKEVSERAEYVGSA 136
Query 113 LDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGE 172
L G +P + +G+ ++ R +W +++ +V VPLYDTLG
Sbjct 137 LLHR-----------GFKSSPD--QFVGIFAQNRPEWTIVELGCYTYSMVAVPLYDTLGA 183
Query 173 PSVKHIVKLTKMEVLAAE 190
++ +I+ + V+ +
Sbjct 184 EAITYIINKADLTVVFCD 201
> ath:AT1G77590 LACS9; LACS9 (LONG CHAIN ACYL-COA SYNTHETASE 9);
long-chain-fatty-acid-CoA ligase (EC:6.2.1.3); K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=691
Score = 43.5 bits (101), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 39/178 (21%), Positives = 73/178 (41%), Gaps = 24/178 (13%)
Query 47 EPAIPQFPHIKSTYPLLKEAARIFGRNLYVGERQ----PIKDSNSQTT-----LSDFKYF 97
EP + HI + L + + +++G R+ I+ S T L D+++
Sbjct 48 EPVSSHWEHISTLPELFEISCNAHSDRVFLGTRKLISREIETSEDGKTFEKLHLGDYEWL 107
Query 98 TYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAAN 157
T+ TL V L V + H EE RV + + TR +W +
Sbjct 108 TFGKTLEAVCDFASGL-----VQIGHKTEE---------RV-AIFADTREEWFISLQGCF 152
Query 158 YRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAALQLKKEGFPLKAVISFDE 215
R + V +Y +LGE ++ H + T++ + +L + + ++ +K VI D+
Sbjct 153 RRNVTVVTIYSSLGEEALCHSLNETEVTTVICGSKELKKLMDISQQLETVKRVICMDD 210
> dre:445175 zgc:101071 (EC:6.2.1.3); K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=697
Score = 43.1 bits (100), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 22/97 (22%), Positives = 49/97 (50%), Gaps = 13/97 (13%)
Query 94 FKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLD 153
+++ +Y++ + E LG A + +G + + P +G+ S+ R +W + +
Sbjct 118 YEWLSYSEVIERAEHLGSA-----------FLHKGHSKSGDPY--IGIFSQNRPEWTIAE 164
Query 154 FAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAE 190
A +++VPLYDTLG ++ +I+ T + + +
Sbjct 165 LACYTYSLISVPLYDTLGTEAISYILDKTCISTVVCD 201
> ath:AT1G64400 long-chain-fatty-acid--CoA ligase, putative /
long-chain acyl-CoA synthetase, putative; K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=665
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 35/125 (28%), Positives = 60/125 (48%), Gaps = 16/125 (12%)
Query 94 FKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLD 153
+ + TY + +V LG ++ + +GV D+ G+ A P +W +
Sbjct 77 YVWQTYKEVHNVVIKLGNSI-RTIGVGKG--DKCGIYGANSP------------EWIISM 121
Query 154 FAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAALQLK-KEGFPLKAVIS 212
A N G+ VPLYDTLG +++ I+ ++ + AE +K+ L+ K LK ++S
Sbjct 122 EACNAHGLYCVPLYDTLGAGAIEFIICHAEVSLAFAEENKISELLKTAPKSTKYLKYIVS 181
Query 213 FDEPT 217
F E T
Sbjct 182 FGEVT 186
> hsa:23305 ACSL6, ACS2, FACL6, FLJ16173, KIAA0837, LACS_6, LACS2,
LACS5; acyl-CoA synthetase long-chain family member 6 (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=722
Score = 42.0 bits (97), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 36/162 (22%), Positives = 69/162 (42%), Gaps = 27/162 (16%)
Query 57 KSTYPLLKEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQE 116
++ Y + + I G +G R+P + +++ +Y + E LG L Q
Sbjct 115 RTMYQVFRRGLSISGNGPCLGFRKPKQP---------YQWLSYQEVADRAEFLGSGLLQH 165
Query 117 VGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVK 176
N + +G+ ++ R +W +++ A +V VPLYDTLG +++
Sbjct 166 -------------NCKACTDQFIGVFAQNRPEWIIVELACYTYSMVVVPLYDTLGPGAIR 212
Query 177 HIVKLTKMEVLAAEGSKLDAALQL----KKEGFPLKAVISFD 214
+I+ + + + + A L L +KE LK +I D
Sbjct 213 YIINTADISTVIVDKPQ-KAVLLLEHVERKETPGLKLIILMD 253
> tgo:TGME49_110150 long-chain-fatty-acid-CoA ligase, putative
(EC:6.2.1.3); K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=866
Score = 42.0 bits (97), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 37/148 (25%), Positives = 65/148 (43%), Gaps = 7/148 (4%)
Query 69 IFGRNLYVGERQPIKDSNSQTTLSD-FKYFTYADTLRMVEALGRALD---QEVGVPLSHY 124
+ G + + G +P ++D F Y TYA V ALG L ++ G+ L +
Sbjct 174 LVGVSAHAGPEKPCIGERRSAAIADGFDYTTYAQVDETVRALGSGLADLMRKDGIKLETF 233
Query 125 DEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAAN-YRGIVTVPLYDTLGEPSVKHIVKLTK 183
+E N +G+ ++R +W D A + Y + +V L+ T VK I + +K
Sbjct 234 PDEERNDG--KFANVGILLQSRKEWLFTDLAVSAYPPLTSVTLHYTFPAEHVKSIAEQSK 291
Query 184 MEVLAAEGSKLDAALQLKKEGFPLKAVI 211
+ + + KL +LK LK ++
Sbjct 292 LSCIVTDVDKLKLLAELKPSLGELKTIV 319
> mmu:216739 Acsl6, A330035H04Rik, AW050338, Facl6, LACS, Lacsl,
MGC36650, mKIAA0837; acyl-CoA synthetase long-chain family
member 6 (EC:6.2.1.3); K01897 long-chain acyl-CoA synthetase
[EC:6.2.1.3]
Length=722
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/124 (23%), Positives = 55/124 (44%), Gaps = 22/124 (17%)
Query 57 KSTYPLLKEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQE 116
++ Y + + I G +G R+P + +++ +Y + + E LG L
Sbjct 115 RTMYQVFRRGLSISGNGPCLGFRKPEQP---------YQWLSYQEVAKRAEFLGSGL--- 162
Query 117 VGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVK 176
L H + G + +G+ ++ R +W + + A +V VPLYDTLG S+
Sbjct 163 ----LQHDCKVGTE------QFVGVFAQNRPEWIIAELACYTYSMVVVPLYDTLGPGSIS 212
Query 177 HIVK 180
+I+
Sbjct 213 YIIN 216
> ath:AT2G47240 long-chain-fatty-acid--CoA ligase family protein
/ long-chain acyl-CoA synthetase family protein (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=660
Score = 41.2 bits (95), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 48/209 (22%), Positives = 86/209 (41%), Gaps = 33/209 (15%)
Query 8 VKTFKGKYALPIEGTETADSSA-VYRAVPDPSNPTQTMLGEPAIPQF-PHIKSTYPLLKE 65
+K+F K ++G + S VYR +L E P I + + + +
Sbjct 1 MKSFAAKVEEGVKGIDGKPSVGPVYR----------NLLSEKGFPPIDSEITTAWDIFSK 50
Query 66 AARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYD 125
+ F N +G R+ + + + + + TY + V +G AL P S
Sbjct 51 SVEKFPDNNMLGWRRIVDEK-----VGPYMWKTYKEVYEEVLQIGSALRAAGAEPGSRVG 105
Query 126 EEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKME 185
G+N P+ W + A ++ VPLYDTLG +V +IV+ +++
Sbjct 106 IYGVN-CPQ--------------WIIAMEACAAHTLICVPLYDTLGSGAVDYIVEHAEID 150
Query 186 VLAAEGSKLDAALQLK-KEGFPLKAVISF 213
+ + +K+ L+ K LKA++SF
Sbjct 151 FVFVQDTKIKGLLEPDCKCAKRLKAIVSF 179
> cel:R09E10.4 hypothetical protein; K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=339
Score = 41.2 bits (95), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 42/79 (53%), Gaps = 5/79 (6%)
Query 139 LGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAAL 198
+G+ S+ R +W L D A + V+VPLYDT+ + +I L ++ ++ A+ D
Sbjct 142 IGIYSKNRPEWILSDMAIHNFSNVSVPLYDTITNDDMHYITNLCEISLIFADSE--DKTT 199
Query 199 QLKKEG---FPLKAVISFD 214
QL ++ LK ++ FD
Sbjct 200 QLVRDKSYLSNLKYIVQFD 218
> pfa:PFL0035c ACS7; acyl-CoA synthetase, PfACS7; K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=926
Score = 41.2 bits (95), Expect = 0.003, Method: Composition-based stats.
Identities = 34/124 (27%), Positives = 55/124 (44%), Gaps = 5/124 (4%)
Query 96 YFTYADTLRMVEALGRALD--QEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLD 153
Y TY + + V + +L+ + GVP Y+EE N R+LGL +W + D
Sbjct 92 YITYGNFFKKVLSFSHSLNTYEGTGVPEKIYNEEKNN---GKFRLLGLYGNNSTNWLITD 148
Query 154 FAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAALQLKKEGFPLKAVISF 213
A G+ T+ ++ + I+ TK+E L + ++ L K E LK +I
Sbjct 149 CACMISGVTTLVMHSKFSIDIIIDILNNTKLEWLCLDLDLVEGLLCRKNELPYLKKLIIL 208
Query 214 DEPT 217
D T
Sbjct 209 DNLT 212
> dre:566863 fa04h02, wu:fc05h07; wu:fa04h02; K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=633
Score = 40.8 bits (94), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 30/134 (22%), Positives = 58/134 (43%), Gaps = 22/134 (16%)
Query 57 KSTYPLLKEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQE 116
K+ Y + + I G ++G R P + + + +Y + E +G L
Sbjct 25 KTMYEVFQRGLHITGDGPFLGSRLPNQP---------YTWLSYREVSTRAEYVGSGL--- 72
Query 117 VGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVK 176
LS +G P L +G+ ++ R +W + + A +V VPLYDTLG +++
Sbjct 73 ----LS----QGCQPNTDQL--IGVFAQNRPEWIISELACYTYSMVVVPLYDTLGADAIR 122
Query 177 HIVKLTKMEVLAAE 190
I+ ++ + +
Sbjct 123 FIINTAEISTVICD 136
> cel:Y65B4BL.5 acs-13; fatty Acid CoA Synthetase family member
(acs-13); K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=719
Score = 40.4 bits (93), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 37/153 (24%), Positives = 72/153 (47%), Gaps = 26/153 (16%)
Query 71 GRNLYVGERQPIKDSNSQTTL------SD----FKYFTYADTLRMVEALGRALDQEVGVP 120
R LY G R+ + SN+ L SD + + +Y L + + A +E+GVP
Sbjct 108 ARTLYQGVRRGARLSNNGPMLGRRVKQSDGSIPYVWESYNTILERADNVSVAF-RELGVP 166
Query 121 LSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAA-NYRGIVTVPLYDTLGEPSVKHIV 179
+ + +G+ S+ R +W + +FA NY ++ VP+Y+TLG + I+
Sbjct 167 TGNAEN------------IGIYSKNRAEWIITEFATYNYSNVI-VPIYETLGSEASIFIL 213
Query 180 KLTKMEVLAAEG-SKLDAALQLKKEGFPLKAVI 211
+++++ + SK L+ K++ L ++
Sbjct 214 NQAEIKIVVCDDISKATGLLKFKEQCPSLSTLV 246
> hsa:51703 ACSL5, ACS2, ACS5, FACL5; acyl-CoA synthetase long-chain
family member 5 (EC:6.2.1.3); K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=739
Score = 40.0 bits (92), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 29/127 (22%), Positives = 52/127 (40%), Gaps = 15/127 (11%)
Query 64 KEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLSH 123
K +F R L V + P +++ +Y E LG L +
Sbjct 131 KTMYEVFQRGLAVSDNGPCLGYRKPN--QPYRWLSYKQVSDRAEYLGSCLLHK------- 181
Query 124 YDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTK 183
G +P + +G+ ++ R +W + + A +V VPLYDTLG ++ HIV
Sbjct 182 ----GYKSSPD--QFVGIFAQNRPEWIISELACYTYSMVAVPLYDTLGPEAIVHIVNKAD 235
Query 184 MEVLAAE 190
+ ++ +
Sbjct 236 IAMVICD 242
> pfa:PF14_0761 ACS1a; acyl-CoA synthetase; K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=820
Score = 38.9 bits (89), Expect = 0.012, Method: Composition-based stats.
Identities = 30/121 (24%), Positives = 55/121 (45%), Gaps = 5/121 (4%)
Query 96 YFTYADTLRMVEALGRALDQEVGV--PLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLD 153
Y TY LR + + +L++ G+ P Y+EE N ++LGL ++W + D
Sbjct 93 YMTYDTFLRKILSFNNSLNKYDGINIPEKIYNEEMNN---GKFKLLGLYGSNSINWLVAD 149
Query 154 FAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAALQLKKEGFPLKAVISF 213
A G+ T+ ++ + I+ TK+E L + ++ ++ E LK +I
Sbjct 150 LGAMLSGVTTLVMHSKFSMDVIVGILNETKLEWLCLDLDLVEGLMERINELPHLKNLIIL 209
Query 214 D 214
D
Sbjct 210 D 210
> cel:F37C12.7 acs-16; fatty Acid CoA Synthetase family member
(acs-16); K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=731
Score = 38.9 bits (89), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 40/189 (21%), Positives = 75/189 (39%), Gaps = 24/189 (12%)
Query 34 VPDPSNPTQTMLGEPAIPQFPHIKSTYPLLKEAARIFGRNLYVGERQPIKDSNSQT---- 89
V D P + + G A FP + + ++ ++ + +G RQ I+ +
Sbjct 71 VDDRDGPIRNIQGH-ASETFPGKDTVDKVWRKCVELYDESPCLGTRQLIEVHEDKQPGGR 129
Query 90 -----TLSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSR 144
L ++ + +Y + V L + L+H ++ NP + + +
Sbjct 130 VFEKWHLGEYVWQSYKEVETQVARLAAGIKD-----LAHEEQ---NPK------VVIFAE 175
Query 145 TRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAALQLKKEG 204
TR DW + A + V +Y TLGE ++ H + T+ +L L + L K+
Sbjct 176 TRADWLITALACFRANVTIVTVYATLGEDAIAHAIGETEATILVTSSELLPKIVTLGKKC 235
Query 205 FPLKAVISF 213
LK +I F
Sbjct 236 PTLKTLIYF 244
> ath:AT1G49430 LACS2; LACS2 (LONG-CHAIN ACYL-COA SYNTHETASE 2);
long-chain-fatty-acid-CoA ligase (EC:6.2.1.3); K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=665
Score = 38.9 bits (89), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 33/161 (20%), Positives = 66/161 (40%), Gaps = 21/161 (13%)
Query 56 IKSTYPLLKEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQ 115
I S + EA + + +G+R + + + + + TY + +G A+
Sbjct 44 IDSPWQFFSEAVKKYPNEQMLGQR-----VTTDSKVGPYTWITYKEAHDAAIRIGSAIRS 98
Query 116 EVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSV 175
G++P G+ +W + A +GI VPLYD+LG +V
Sbjct 99 R-----------GVDPG----HCCGIYGANCPEWIIAMEACMSQGITYVPLYDSLGVNAV 143
Query 176 KHIVKLTKMEVLAAEGSKLDAALQLKKEGFP-LKAVISFDE 215
+ I+ ++ ++ + + + L +K LK ++SF E
Sbjct 144 EFIINHAEVSLVFVQEKTVSSILSCQKGCSSNLKTIVSFGE 184
> sce:YER015W FAA2, FAM1; Faa2p (EC:6.2.1.3); K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=744
Score = 38.9 bits (89), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 35/163 (21%), Positives = 71/163 (43%), Gaps = 9/163 (5%)
Query 55 HIKSTYPLLKEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALD 114
++++ Y +AR + + +G R PI D + T F++ +Y+ + +G +
Sbjct 91 NLRTAYDHFMFSARRWPQRDCLGSR-PI-DKATGTWEETFRFESYSTVSKRCHNIGSGI- 147
Query 115 QEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPS 174
LS + + P V+ + S +W L D A + LY+TLG +
Sbjct 148 ------LSLVNTKRKRPLEANDFVVAILSHNNPEWILTDLACQAYSLTNTALYETLGPNT 201
Query 175 VKHIVKLTKMEVLAAEGSKLDAALQLKKEGFPLKAVISFDEPT 217
++I+ LT+ +L S + L++ + + ++ DE T
Sbjct 202 SEYILNLTEAPILIFAKSNMYHVLKMVPDMKFVNTLVCMDELT 244
> pfa:PFC0050c PfACS2; acyl-CoA synthetase, PfACS2 (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=819
Score = 38.1 bits (87), Expect = 0.023, Method: Composition-based stats.
Identities = 28/121 (23%), Positives = 54/121 (44%), Gaps = 5/121 (4%)
Query 96 YFTYADTLRMVEALGRALDQEVG--VPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLD 153
Y TY + + V + +L+ G +P Y+EE N ++LG+ ++W + D
Sbjct 92 YMTYGNFFKKVLSFSNSLNTYEGSNIPEKIYNEEMNN---GKFKLLGIYGSNSINWLVAD 148
Query 154 FAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAALQLKKEGFPLKAVISF 213
A G+ T+ ++ + I+ TK+E L + ++ + + E LK +I
Sbjct 149 LGAMLSGVTTLVMHSKFSMDVIVDILNETKLEWLCLDLDLVECLMARRNELPHLKNLIIL 208
Query 214 D 214
D
Sbjct 209 D 209
> sce:YOR317W FAA1; Faa1p (EC:6.2.1.3); K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=700
Score = 37.7 bits (86), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 31/124 (25%), Positives = 55/124 (44%), Gaps = 13/124 (10%)
Query 91 LSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWR 150
LS + Y ++ ++ +GR L V + L D++ L+ L + T W
Sbjct 94 LSHYHYNSFDQLTDIMHEIGRGL---VKIGLKPNDDDKLH----------LYAATSHKWM 140
Query 151 LLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAALQLKKEGFPLKAV 210
+ A +GI V YDTLGE + H + T + + + S L + ++ + +K +
Sbjct 141 KMFLGAQSQGIPVVTAYDTLGEKGLIHSLVQTGSKAIFTDNSLLPSLIKPVQAAQDVKYI 200
Query 211 ISFD 214
I FD
Sbjct 201 IHFD 204
> cel:R09E10.3 hypothetical protein; K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=700
Score = 37.4 bits (85), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 21/80 (26%), Positives = 41/80 (51%), Gaps = 1/80 (1%)
Query 139 LGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGS-KLDAA 197
+G+ S R +W L + A + V+VPLYDT+ + +I L ++ ++ + K
Sbjct 144 IGIYSNNRPEWILSEMAIHNFSNVSVPLYDTITNDDMHYITNLCEISLMFVDAEIKTKQL 203
Query 198 LQLKKEGFPLKAVISFDEPT 217
++ K LK ++ F+E +
Sbjct 204 IRDKSYLSSLKYIVQFNECS 223
> cel:R07C3.4 hypothetical protein; K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=731
Score = 37.4 bits (85), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 22/81 (27%), Positives = 41/81 (50%), Gaps = 6/81 (7%)
Query 139 LGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAAL 198
+G+ S+ R +W L D A + V+VP+YD + + +I L ++ ++ + D
Sbjct 140 IGIYSKNRPEWVLSDMAIHNFSNVSVPIYDNIPNEDMHYITNLCEIPLMFVDSE--DKTT 197
Query 199 QLKKE----GFPLKAVISFDE 215
QL K+ LK ++ FD+
Sbjct 198 QLIKDKTYLSKSLKYIVQFDK 218
> ath:AT4G23850 long-chain-fatty-acid--CoA ligase / long-chain
acyl-CoA synthetase; K01897 long-chain acyl-CoA synthetase
[EC:6.2.1.3]
Length=666
Score = 37.4 bits (85), Expect = 0.043, Method: Composition-based stats.
Identities = 50/214 (23%), Positives = 86/214 (40%), Gaps = 51/214 (23%)
Query 14 KYALPIE-GTETADS----SAVYRAV------PDPSNPTQTMLGEPAIPQFPHIKSTYPL 62
KY +E G E +D VYR++ PDP + S + +
Sbjct 6 KYIFQVEEGKEGSDGRPSVGPVYRSIFAKDGFPDP---------------IEGMDSCWDV 50
Query 63 LKEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLS 122
+ + + N +G R+ + + + + TY + +V LG +L + VGV
Sbjct 51 FRMSVEKYPNNPMLGRREIVDGKPGK-----YVWQTYQEVYDIVMKLGNSL-RSVGVK-- 102
Query 123 HYDEE--GLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVK 180
DE G+ A P +W + A N G+ VPLYDTLG +V+ I+
Sbjct 103 --DEAKCGIYGANSP------------EWIISMEACNAHGLYCVPLYDTLGADAVEFIIS 148
Query 181 LTKMEVLAAEGSKLDAALQLKKEGFP-LKAVISF 213
+++ ++ E K+ + +K V+SF
Sbjct 149 HSEVSIVFVEEKKISELFKTCPNSTEYMKTVVSF 182
> dre:559656 acsl3b, acsl3, fb34c03, im:7155129, si:dkey-20f20.5,
wu:fa07a08, wu:fb34c03; acyl-CoA synthetase long-chain family
member 3b; K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=711
Score = 37.0 bits (84), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 34/142 (23%), Positives = 54/142 (38%), Gaps = 17/142 (11%)
Query 78 ERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPLR 137
E QP + L D+ + +Y DT + + G L KPL
Sbjct 107 EIQPNGKVFKKVILGDYNWLSYQDTFHLAQRFGSGLAA---------------LGQKPLC 151
Query 138 VLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAA 197
+ + TR +W + A V LY TLG ++ H + T++ + L +
Sbjct 152 NIAIFCETRAEWVIAAQACFMYNFPLVTLYSTLGGSAIAHGLNETEVTHIVTSKDLLQSR 211
Query 198 LQLKKEGFP-LKAVISFD-EPT 217
L+ P LK +I D +PT
Sbjct 212 LKAILLEVPRLKHIIVVDTKPT 233
> dre:555308 acsl1, MGC110081, zgc:110081; acyl-CoA synthetase
long-chain family member 1 (EC:6.2.1.3); K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=697
Score = 36.6 bits (83), Expect = 0.064, Method: Compositional matrix adjust.
Identities = 13/43 (30%), Positives = 27/43 (62%), Gaps = 0/43 (0%)
Query 137 RVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIV 179
+ +G+ ++ R +W + + A +V VPLYDTLG ++ +++
Sbjct 148 KYIGIFAQNRPEWTISELACYTYSLVAVPLYDTLGTEAISYVI 190
> ath:AT4G11030 long-chain-fatty-acid--CoA ligase, putative /
long-chain acyl-CoA synthetase, putative (EC:6.2.1.3); K01897
long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=666
Score = 35.4 bits (80), Expect = 0.13, Method: Composition-based stats.
Identities = 40/166 (24%), Positives = 71/166 (42%), Gaps = 25/166 (15%)
Query 53 FPH----IKSTYPLLKEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEA 108
FP+ I+S + + + A + N +G R+ SN + + + TY + +V
Sbjct 37 FPNPIDGIQSCWDIFRTAVEKYPNNRMLGRREI---SNGKA--GKYVWKTYKEVYDIVIK 91
Query 109 LGRALDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYD 168
LG +L + G+ +EG G+ +W + A N G+ VPLYD
Sbjct 92 LGNSL-RSCGI------KEG--------EKCGIYGINCCEWIISMEACNAHGLYCVPLYD 136
Query 169 TLGEPSVKHIVKLTKMEVLAAEGSKLDAALQLKKEGFP-LKAVISF 213
TLG +V+ I+ ++ + E K+ + +K V+SF
Sbjct 137 TLGAGAVEFIISHAEVSIAFVEEKKIPELFKTCPNSTKYMKTVVSF 182
> mmu:433256 Acsl5, 1700030F05Rik, ACS2, ACS5, Facl5; acyl-CoA
synthetase long-chain family member 5 (EC:6.2.1.3); K01897
long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=683
Score = 35.4 bits (80), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 14/54 (25%), Positives = 30/54 (55%), Gaps = 0/54 (0%)
Query 137 RVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAE 190
+ +G+ ++ R +W + + A +V VPLYDTLG ++ ++ + V+ +
Sbjct 133 QFVGIFAQNRPEWVISELACYTYSMVAVPLYDTLGTEAIIFVINRADIPVVICD 186
> pfa:PF07_0129 PfACS5; acyl-coA synthetase, PfACS5 (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=811
Score = 35.4 bits (80), Expect = 0.15, Method: Composition-based stats.
Identities = 29/120 (24%), Positives = 50/120 (41%), Gaps = 2/120 (1%)
Query 96 YFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFA 155
Y TY D + V L+ G + + R+LGL ++W D A
Sbjct 92 YLTYNDFFKKVLCFSNTLNTYEGKGIEEKMYKKEEKNNGKFRLLGLYGSNSINWIAADMA 151
Query 156 ANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAALQLKKEGFP-LKAVISFD 214
+ G+ T+ ++ + I+K T++E L + ++ L + E FP LK +I D
Sbjct 152 SMLSGVTTLVMHSKFSLDVIVDILKETELEWLCLDLELVEGLLAHRNE-FPHLKNLIILD 210
> dre:567157 acsl3a, si:dkeyp-109h9.2; acyl-CoA synthetase long-chain
family member 3a (EC:6.2.1.3); K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=713
Score = 35.4 bits (80), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 20/90 (22%), Positives = 35/90 (38%), Gaps = 15/90 (16%)
Query 88 QTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRM 147
+ L ++++ +Y + V A G L KP++ + + TR
Sbjct 119 KVILGEYRWMSYEQVFKEVNAFGSGLAA---------------LGQKPMKNIAIFCETRA 163
Query 148 DWRLLDFAANYRGIVTVPLYDTLGEPSVKH 177
+W + A V LY TLG P++ H
Sbjct 164 EWIIAAQACFMYNFPLVTLYSTLGGPAIAH 193
> pfa:PF14_0751 ACS1b; acyl-CoA synthetase, PfACS1b; K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=770
Score = 35.0 bits (79), Expect = 0.23, Method: Composition-based stats.
Identities = 28/119 (23%), Positives = 52/119 (43%), Gaps = 1/119 (0%)
Query 96 YFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFA 155
Y TY+D + V + L G + + + K ++LGL ++W + D
Sbjct 92 YITYSDFFKKVLSFSHNLCTYEGKGIESQSYKEIRNDGK-FKLLGLYGSNSINWLVSDLG 150
Query 156 ANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAALQLKKEGFPLKAVISFD 214
+ G+ T+ L+ + I+ TK+E + + ++ L+ KK+ LK VI D
Sbjct 151 SMISGVTTLVLHSKFSIDVIVGILNETKLEWICVDLDLVEGILERKKDLPYLKNVIILD 209
> pfa:PFD0085c PfACS6; acyl-CoA synthetase, PfACS6 (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=971
Score = 34.3 bits (77), Expect = 0.30, Method: Composition-based stats.
Identities = 29/121 (23%), Positives = 51/121 (42%), Gaps = 5/121 (4%)
Query 96 YFTYADTLRMVEALGRALDQE--VGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLD 153
+ TY V + +LD G+ YDE+ N ++LGL ++W + D
Sbjct 92 FMTYGTFYEKVCSFSHSLDTYGGKGIEARKYDEDKNNGM---FKLLGLYGSNSINWLVTD 148
Query 154 FAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAALQLKKEGFPLKAVISF 213
G+ T+ ++ + I+K T++E L + ++ L KE LK +I
Sbjct 149 MGCMMSGVTTIVMHSKFSIDLIVDILKRTQLEWLCIDLDLVEGLLCHIKELPHLKKLIIL 208
Query 214 D 214
D
Sbjct 209 D 209
Lambda K H
0.317 0.135 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7009334380
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40