bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0167_orf1
Length=181
Score E
Sequences producing significant alignments: (Bits) Value
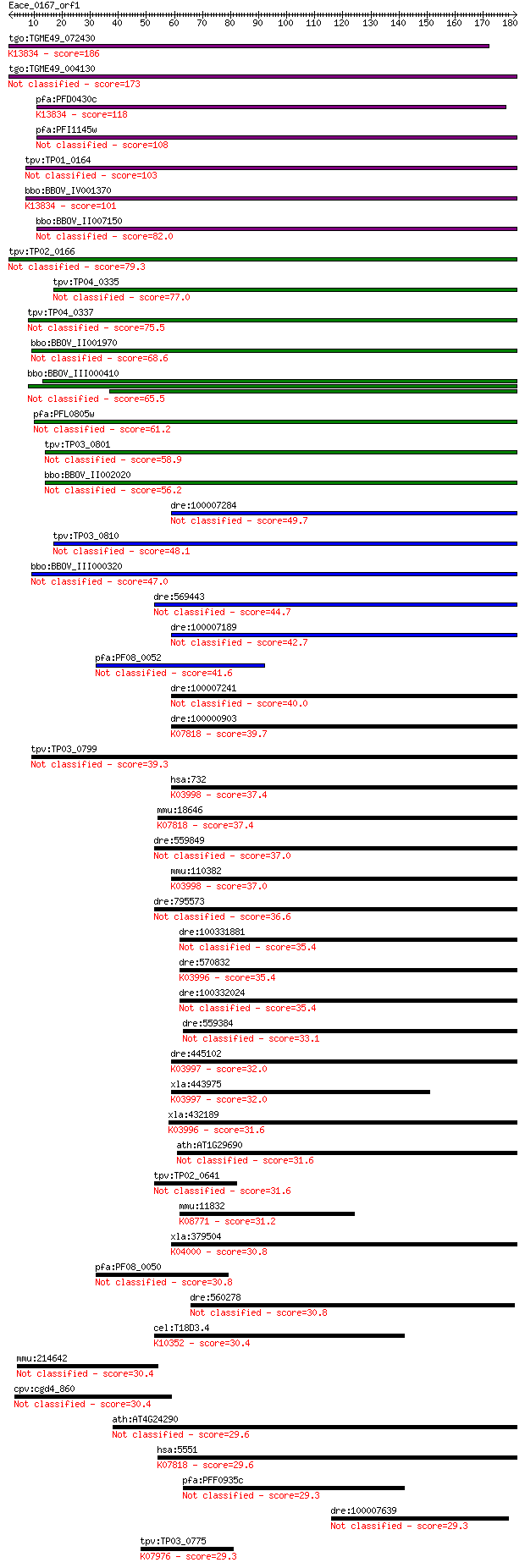
tgo:TGME49_072430 membrane-attack complex / perforin domain-co... 186 3e-47
tgo:TGME49_004130 membrane-attack complex / perforin domain-co... 173 3e-43
pfa:PFD0430c MAC/Perforin, putative; K13834 sporozoite microne... 118 1e-26
pfa:PFI1145w MAC/Perforin, putative 108 9e-24
tpv:TP01_0164 hypothetical protein 103 4e-22
bbo:BBOV_IV001370 21.m02755; MAC/perforin domain containing pr... 101 1e-21
bbo:BBOV_II007150 18.m06592; mac/perforin domain containing pr... 82.0 1e-15
tpv:TP02_0166 hypothetical protein 79.3 6e-15
tpv:TP04_0335 hypothetical protein 77.0 3e-14
tpv:TP04_0337 hypothetical protein 75.5 9e-14
bbo:BBOV_II001970 18.m09950; mac/perforin domain containing me... 68.6 1e-11
bbo:BBOV_III000410 hypothetical protein 65.5 9e-11
pfa:PFL0805w MAC/Perforin, putative 61.2 2e-09
tpv:TP03_0801 hypothetical protein 58.9 9e-09
bbo:BBOV_II002020 18.m06160; mac/perforin domain containing pr... 56.2 6e-08
dre:100007284 Perforin-1-like 49.7 5e-06
tpv:TP03_0810 hypothetical protein 48.1 2e-05
bbo:BBOV_III000320 17.m10445; hypothetical protein 47.0 3e-05
dre:569443 Perforin-1-like 44.7 2e-04
dre:100007189 Perforin-1-like 42.7 7e-04
pfa:PF08_0052 perforin like protein 5 41.6
dre:100007241 Perforin-1-like 40.0 0.004
dre:100000903 Perforin-1-like; K07818 perforin 1 39.7
tpv:TP03_0799 hypothetical protein 39.3 0.007
hsa:732 C8B, MGC163447; complement component 8, beta polypepti... 37.4 0.031
mmu:18646 Prf1, Pfn, Pfp, Prf-1; perforin 1 (pore forming prot... 37.4 0.031
dre:559849 similar to perforin 1 (pore forming protein) 37.0 0.036
mmu:110382 C8b, 4930439B20Rik, AI595927; complement component ... 37.0 0.041
dre:795573 MGC163021; zgc:163021 36.6 0.044
dre:100331881 hypothetical protein LOC100331881 35.4 0.097
dre:570832 c7, cb349, wu:fa01f03; complement component 7; K039... 35.4 0.10
dre:100332024 hypothetical protein LOC100332024 35.4 0.10
dre:559384 si:ch211-103n10.4 33.1 0.57
dre:445102 c8a, zgc:92465; complement component 8, alpha polyp... 32.0 1.1
xla:443975 c8a, MGC80388; complement component 8, alpha polype... 32.0 1.1
xla:432189 c7, MGC82368; complement component 7; K03996 comple... 31.6 1.4
ath:AT1G29690 CAD1; CAD1 (constitutively activated cell death 1) 31.6 1.4
tpv:TP02_0641 hypothetical protein 31.6 1.5
mmu:11832 Aqp7, AQP7L, AQPap; aquaporin 7; K08771 aquaporin-7 31.2 2.2
xla:379504 c9, MGC64276; complement component 9; K04000 comple... 30.8 2.4
pfa:PF08_0050 MAC/Perforin, putative 30.8 2.4
dre:560278 hypothetical LOC560278 30.8 2.7
cel:T18D3.4 myo-2; MYOsin heavy chain structural genes family ... 30.4 3.1
mmu:214642 A430107O13Rik, 6720481P07, AI552584; RIKEN cDNA A43... 30.4 3.4
cpv:cgd4_860 possible WWE domain 30.4 3.7
ath:AT4G24290 hypothetical protein 29.6 6.2
hsa:5551 PRF1, FLH2, HPLH2, MGC65093, P1, PFN1, PFP; perforin ... 29.6 6.3
pfa:PFF0935c conserved Plasmodium protein, unknown function 29.3 7.2
dre:100007639 pancreatic secretory granule membrane major glyc... 29.3 7.3
tpv:TP03_0775 small GTP-binding protein Rab1; K07976 Rab famil... 29.3 8.0
> tgo:TGME49_072430 membrane-attack complex / perforin domain-containing
protein ; K13834 sporozoite microneme protein 2
Length=854
Score = 186 bits (473), Expect = 3e-47, Method: Compositional matrix adjust.
Identities = 84/171 (49%), Positives = 118/171 (69%), Gaps = 0/171 (0%)
Query 1 AGLAQTDNFNWNYTLAFNEAVAGLPPTFDGADEGKGCSPEVWRENTKDKLCADSTVKTWM 60
AGLAQTD+F WNYTLAF++AVA LP TFDG + CS + WR + C + + WM
Sbjct 363 AGLAQTDSFKWNYTLAFDDAVAHLPVTFDGNERDTPCSVQQWRADHMADGCQQTNIPIWM 422
Query 61 NFIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGSVGGSTNV 120
FI+QFGTHY RL+AGGK+T+Q+TM +SD+ AL G +VK+ +K G S G S+ V
Sbjct 423 AFIEQFGTHYTARLYAGGKMTYQVTMKSSDVKALKKKGVDVKAEVKLMLGGFSAGASSQV 482
Query 121 EKDEEQKEQLKHFDYQTETLIMGGRVPRDVSDPDSLAQWSDSVEELPMPVK 171
+ +++ QL+ + + E L++GG+ P DVSDP ++A W++SV+ LPMPVK
Sbjct 483 KTNQDSASQLRSLNVEKEALVIGGKPPADVSDPKAIAAWANSVDALPMPVK 533
> tgo:TGME49_004130 membrane-attack complex / perforin domain-containing
protein
Length=1054
Score = 173 bits (439), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 81/181 (44%), Positives = 117/181 (64%), Gaps = 0/181 (0%)
Query 1 AGLAQTDNFNWNYTLAFNEAVAGLPPTFDGADEGKGCSPEVWRENTKDKLCADSTVKTWM 60
AG+AQ+++F WN T+AF V+ LP FD + CS E WR++ + C + V W+
Sbjct 500 AGVAQSNHFKWNVTIAFAAGVSQLPDVFDAHNPECACSAEQWRQDQNAEACTKTNVPIWI 559
Query 61 NFIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGSVGGSTNV 120
+FI+QFGTH++VRLFAGGK+T+Q+T S++ + ++G +VK+ LK G S G
Sbjct 560 SFIEQFGTHFLVRLFAGGKMTYQVTAKRSEVEKMRNMGIDVKTQLKMQLGGVSGGAGQGT 619
Query 121 EKDEEQKEQLKHFDYQTETLIMGGRVPRDVSDPDSLAQWSDSVEELPMPVKLTLQPLEYL 180
+ Q + Q ETL++GGR P +VSDP +LA W+D+VEELPMPVK +QPL +L
Sbjct 620 SSKKNQSSSEYQMNVQKETLVIGGRPPGNVSDPAALAAWADTVEELPMPVKFEVQPLYHL 679
Query 181 L 181
L
Sbjct 680 L 680
> pfa:PFD0430c MAC/Perforin, putative; K13834 sporozoite microneme
protein 2
Length=842
Score = 118 bits (295), Expect = 1e-26, Method: Composition-based stats.
Identities = 58/167 (34%), Positives = 89/167 (53%), Gaps = 2/167 (1%)
Query 11 WNYTLAFNEAVAGLPPTFDGADEGKGCSPEVWRENTKDKLCADSTVKTWMNFIDQFGTHY 70
W+ T A+ AV LP F G D+ C +V+ EN C + V WM F D +GTH
Sbjct 381 WDKTTAYKNAVNELPAVFTGLDKESECPSDVYEENKTKSNCEN--VSLWMKFFDIYGTHI 438
Query 71 VVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGSVGGSTNVEKDEEQKEQL 130
+ GGK+T I +S S I + G +VK+ ++A FG GS GGST+V
Sbjct 439 IYESQLGGKITKIINVSTSSIEQMKKNGVSVKAKIQAQFGFGSAGGSTDVNSSNSSANDE 498
Query 131 KHFDYQTETLIMGGRVPRDVSDPDSLAQWSDSVEELPMPVKLTLQPL 177
+ +D + +++GG +DV+ ++L +WS +V PMP+ + L P+
Sbjct 499 QSYDMNEQLIVIGGNPIKDVTKEENLFEWSKTVTNHPMPINIKLTPI 545
> pfa:PFI1145w MAC/Perforin, putative
Length=821
Score = 108 bits (271), Expect = 9e-24, Method: Composition-based stats.
Identities = 62/172 (36%), Positives = 97/172 (56%), Gaps = 2/172 (1%)
Query 11 WNYTLAFNEAVAGLPPTFDGAD-EGKGCSPEVWRENTKDKLCADSTVKTWMNFIDQFGTH 69
W T F A++ LP F+ + +G CS E +R+N K + C S V WM F FGTH
Sbjct 376 WKLTDQFVRAISLLPSHFNSLEKDGTYCSDEEFRDNRKSEKCGKS-VTAWMYFFKNFGTH 434
Query 70 YVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGSVGGSTNVEKDEEQKEQ 129
L GGK+T Q+ +S +D A++ G ++ +++ A FG+ V GSTN E +E +
Sbjct 435 VSTLLHLGGKITQQVKISKNDYKAMTESGLSISASVSAGFGLFKVKGSTNTESNESSNNE 494
Query 130 LKHFDYQTETLIMGGRVPRDVSDPDSLAQWSDSVEELPMPVKLTLQPLEYLL 181
+ ET+I+GG D +DP++ +W++S+ E PMP+K +PL +L
Sbjct 495 SSTSSLEKETVIIGGTTIFDPNDPNNFEKWAESISENPMPIKGEYEPLSRIL 546
> tpv:TP01_0164 hypothetical protein
Length=1182
Score = 103 bits (257), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 58/178 (32%), Positives = 93/178 (52%), Gaps = 8/178 (4%)
Query 7 DNFNWNYTLAFNEAVAGLPPTFDGADEGKGCSPEVWRENTKDKLCADSTVKTWMNFIDQF 66
D + + T F +AV LP FD C+ E ++ N D +CA+ TV WM FI F
Sbjct 719 DYHSLDTTDEFKKAVEALPDKFDS----HSCTIETFKSNEDDSICAE-TVLPWMQFIKMF 773
Query 67 GTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFG---VGSVGGSTNVEKD 123
GTH+ + GGK+THQ+ + SD+ + G NV +A+KA+ V S+ G +
Sbjct 774 GTHFTTIVHLGGKITHQVQIDKSDVLHMQQNGINVDAAVKASISPVMVDSLQGGFASTSE 833
Query 124 EEQKEQLKHFDYQTETLIMGGRVPRDVSDPDSLAQWSDSVEELPMPVKLTLQPLEYLL 181
+ Q + Y + L++GG D + +SL W+ + + PMP+K+ L+ ++ LL
Sbjct 834 KASLSQSNNLKYDKQVLVIGGDGLVDSKNANSLNNWAKELYKRPMPIKIKLESIKSLL 891
> bbo:BBOV_IV001370 21.m02755; MAC/perforin domain containing
protein; K13834 sporozoite microneme protein 2
Length=978
Score = 101 bits (252), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 55/176 (31%), Positives = 93/176 (52%), Gaps = 6/176 (3%)
Query 7 DNFNWNYTLAFNEAVAGLPPTFDGADEGKGCSPEVWRENTKDKLCADSTVKTWMNFIDQF 66
D N T +F V+ LP F + C EV+R + +D+ CA S V+ WM F ++
Sbjct 460 DFLNIEPTESFVYDVSQLPEKFHDGE----CLLEVYRNDPEDEKCA-SVVRPWMKFFQKY 514
Query 67 GTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKA-TFGVGSVGGSTNVEKDEE 125
GTH+ + GGK+T+QI + D+A L G N+ +K+ + VGG + EK EE
Sbjct 515 GTHFTTVIHLGGKVTNQIQIDKKDVAKLQKDGYNIDVMIKSGSISPVKVGGGFSHEKKEE 574
Query 126 QKEQLKHFDYQTETLIMGGRVPRDVSDPDSLAQWSDSVEELPMPVKLTLQPLEYLL 181
+ +++GG VP D +D ++ +W+ S+ PMP+K+ ++ ++ L+
Sbjct 575 SSSNFSSLQTEKLVIVIGGDVPTDGTDKTTMLEWTKSLYRKPMPIKVNIESIKTLI 630
> bbo:BBOV_II007150 18.m06592; mac/perforin domain containing
protein
Length=752
Score = 82.0 bits (201), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 49/171 (28%), Positives = 75/171 (43%), Gaps = 1/171 (0%)
Query 11 WNYTLAFNEAVAGLPPTFDGADEGKGCSPEVWRENTKDKLCADSTVKTWMNFIDQFGTHY 70
W+ T AF A+ GLP TF G C P + + + + C + V WM F FGTH
Sbjct 560 WDTTQAFQAALKGLPKTFQEEGNGLVCHPYDYLVHPRSQACKELGVTAWMQFFTVFGTHV 619
Query 71 VVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGSVGGSTNVEKDEEQKEQL 130
+++ GGK+ I +S A L G +VK+ L V +V S N
Sbjct 620 TTKVYLGGKMLTIIETKSSQEADLKKKGIDVKAELSVQAEVATVDASVNASTSSLHNRDT 679
Query 131 KHFDYQTETLIMGGRVPRDVSDPDSLAQWSDSVEELPMPVKLTLQPLEYLL 181
+ D + ++GG + D + W+ +V E MP+K PL ++
Sbjct 680 ESLDTKKSMFVIGGDIYGD-GNTIVFNDWAATVPENSMPIKAEYTPLAMIM 729
> tpv:TP02_0166 hypothetical protein
Length=812
Score = 79.3 bits (194), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 44/181 (24%), Positives = 86/181 (47%), Gaps = 4/181 (2%)
Query 1 AGLAQTDNFNWNYTLAFNEAVAGLPPTFDGADEGKGCSPEVWRENTKDKLCADSTVKTWM 60
AG++ + W++TL F ++ L F G ++ C P ++RE+ K++ C + + WM
Sbjct 475 AGISTS--LKWDFTLGFQSSLGRLS-DFKGLEKDSICKPFIYREDPKNENCQELGISDWM 531
Query 61 NFIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGSVGGSTNV 120
+ FGTH +++ GGK+ + + S LS G +V++ L A + + V
Sbjct 532 ELFNTFGTHVATKIYLGGKIFTTLEIKKSQEKKLSDQGLDVRAILSAKIKDTDIDSNVEV 591
Query 121 EKDEEQKEQLKHFDYQTETLIMGGRVPRDVSDPDSLAQWSDSVEELPMPVKLTLQPLEYL 180
+ + D + T ++GG + + A+W+ SV + MP+K P+ +
Sbjct 592 STIKSKNAGDFLLDTKKSTFVLGGDIYGHGKTIE-FAEWARSVADHAMPIKAEFTPISHF 650
Query 181 L 181
+
Sbjct 651 I 651
> tpv:TP04_0335 hypothetical protein
Length=441
Score = 77.0 bits (188), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 48/165 (29%), Positives = 85/165 (51%), Gaps = 8/165 (4%)
Query 17 FNEAVAGLPPTFDGADEGKGCSPEVWRENTKDKLCADSTVKTWMNFIDQFGTHYVVRLFA 76
FN +V+ LP ++ C+PE ++ K+ C D + WM F +FGTH VV++
Sbjct 196 FNLSVSELPKKIKDYEK---CTPEGYKNRIKE--CKD--LVKWMEFFSEFGTHVVVQVHL 248
Query 77 GGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGSVGGSTNVEKDEEQKEQLKHFDYQ 136
G KLT +++ +S I +L++ G +V +A+ A S + V K EE + + +
Sbjct 249 G-KLTRYLSVPSSIIESLANKGLDVNAAIGAVISGVSANIAVGVSKSEESEIKQLKKSSK 307
Query 137 TETLIMGGRVPRDVSDPDSLAQWSDSVEELPMPVKLTLQPLEYLL 181
+ ++GG P P S+ +W +V PMP+K+ ++ + L
Sbjct 308 LKFSVLGGIHPDRNISPTSIRKWKSTVPRYPMPIKIDVESISTFL 352
> tpv:TP04_0337 hypothetical protein
Length=498
Score = 75.5 bits (184), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 44/174 (25%), Positives = 89/174 (51%), Gaps = 9/174 (5%)
Query 8 NFNWNYTLAFNEAVAGLPPTFDGADEGKGCSPEVWRENTKDKLCADSTVKTWMNFIDQFG 67
N W YT FN ++ LP + K C+ + + N D+ C ++K W+ F + FG
Sbjct 148 NLKWGYTEYFNRTLSRLP--ILSSKVIKNCNIDN-KLNLSDEECK--SIKPWIKFFEVFG 202
Query 68 THYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGSVGGSTNVEKDEEQK 127
TH+ +L GGK+ + +S + L G ++++ ++ G G+V + ++ + +
Sbjct 203 THFNNQLTLGGKINQTMVFDSSTLEELKKKGIDIEAEVRTELGSGNVKLNLDMGGKKSRL 262
Query 128 EQLKHFDYQTETLIMGGRVPRDVSDPDSLAQWSDSVEELPMPVKLTLQPLEYLL 181
+++ Q + ++GG++P D + A W+++V E PMP+ + L+ L+
Sbjct 263 DEIG----QKKMSVLGGKMPNFPMDDNEFAHWAETVAENPMPIGVVSTSLKTLM 312
> bbo:BBOV_II001970 18.m09950; mac/perforin domain containing
membrane protein
Length=559
Score = 68.6 bits (166), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 46/173 (26%), Positives = 78/173 (45%), Gaps = 27/173 (15%)
Query 9 FNWNYTLAFNEAVAGLPPTFDGADEGKGCSPEVWRENTKDKLCADSTVKTWMNFIDQFGT 68
FNW T F+ A+ LP D+ C+ E++++ K K C ++ W++F QFGT
Sbjct 194 FNWETTSDFDIALNELPKEVKSFDQ---CTVELYKK--KSKKCG--SISKWVDFFLQFGT 246
Query 69 HYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGSVGGSTNVEKDEEQKE 128
H + GGK+ +T+ N+ + + G NV A+KA + N + + EQ++
Sbjct 247 HVTTEIQLGGKIIRLLTIPNNAMDSFLKSGLNVDVAVKAVISGALL--EVNEKLNSEQQK 304
Query 129 QLKHFDYQTETLIMGGRVPRDVSDPDSLAQWSDSVEELPMPVKLTLQPLEYLL 181
+K DSL +W +V PMP++ PL+ +
Sbjct 305 AIKEL------------------QDDSLLKWKSTVPIFPMPIRTIYAPLDMFI 339
> bbo:BBOV_III000410 hypothetical protein
Length=1272
Score = 65.5 bits (158), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 50/176 (28%), Positives = 81/176 (46%), Gaps = 20/176 (11%)
Query 13 YTLAFNEAVAGLPPTFDGADEGKGCSPEVWRENTKDKLCADSTVKTWMNFIDQFGTHYVV 72
+ +F A L P F A C+ + N + C + VK WM + FGTH+
Sbjct 1077 FKKSFQNATKELKPNFKKA--ATECTTIRYAINPEYPDCKE--VKPWMQLFEMFGTHFTY 1132
Query 73 RLFAGGKLTH--QITMSNSDIAALSSLG----TNVKSAL-KATFGVGSVGGSTNVEKDEE 125
+ GG+LT Q+ ++ ++S+ T V++ L A+ GVG GST KD
Sbjct 1133 NIKIGGRLTKISQVNLNKKTNTNMNSVNAGATTQVRNELFGASAGVGLSKGST---KDNT 1189
Query 126 QKEQLKHFDYQTETLIMGGRVPRDVSDPDSLAQWSDSVEELPMPVKLTLQPLEYLL 181
+ + + ++GGR +V D + +W +S+ E PMP++ L PL L
Sbjct 1190 ENISFTYVN------VLGGRTIGNVEDENEYLEWINSIPEHPMPIRNQLAPLAKLF 1239
Score = 59.3 bits (142), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 49/188 (26%), Positives = 84/188 (44%), Gaps = 19/188 (10%)
Query 8 NFNWNYTLAFNEAVAGLP----------PTFDGADEGKGCSPEVWRENTKDKLCADSTVK 57
NF NY + N+ +AG+ F+ A + P+ + +T++K +D+ K
Sbjct 226 NFGVNYVMQ-NKEIAGMEDIVKKHMNVTRAFENATKEIESYPDKEKCDTQEKYLSDAECK 284
Query 58 T----WMNFIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGS 113
W F +GTH + R+ GGK+ M N+ A + +N K + VG
Sbjct 285 NYCELWKKFFMNYGTHVISRITMGGKILQMDEMENT---ANENAESNDKKH-NISVNVGV 340
Query 114 VGGSTNVEKDEEQKEQLKHFDYQTETLIMGGRVPRDVSDPDSLAQWSDSVEELPMPVKLT 173
+ GS N+ E++++ K + IMGG V P+ W DS++E MP+ +
Sbjct 341 LSGSFNLNNTNEKQQKEKVKKKNGKFFIMGGDTFIPVDTPEGFRDWVDSIKENSMPINVE 400
Query 174 LQPLEYLL 181
L P+ +
Sbjct 401 LTPMSQFM 408
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 34/146 (23%), Positives = 55/146 (37%), Gaps = 5/146 (3%)
Query 37 CSPEVWRENTKDKLCADSTVKTWMNFIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSS 96
C E + EN D+ C+ + +WM F +GTH R+ GG +SN + +S
Sbjct 543 CPTEKYVENMDDESCS-GCISSWMRFFADYGTHVTRRISMGGIFRRFSNVSNRESKRDAS 601
Query 97 LGTNVKSALKATFGVGSVGGSTNVEKDEEQKEQLKHFDYQTETLIMGGRVPRDVS-DPDS 155
+ F + + + K D T+ G P + S PD
Sbjct 602 KQKTTIKKSSSWFHLVKKKSKKTKSSSSKVSSEGKEHDLVQYTV---GPEPHNESMAPDV 658
Query 156 LAQWSDSVEELPMPVKLTLQPLEYLL 181
W V P+P+ + QPL ++
Sbjct 659 FEDWVRDVALNPVPIDVEYQPLSEIM 684
> pfa:PFL0805w MAC/Perforin, putative
Length=1073
Score = 61.2 bits (147), Expect = 2e-09, Method: Composition-based stats.
Identities = 43/172 (25%), Positives = 75/172 (43%), Gaps = 5/172 (2%)
Query 10 NWNYTLAFNEAVAGLPPTFDGADEGKGCSPEVWRENTKDKLCADSTVKTWMNFIDQFGTH 69
++ ++ F A++ LP FDG E CS E + C + V WM F GTH
Sbjct 604 SYKFSENFKNALSKLPKYFDGLREDSKCSYEYYINKLNSPEC-EENVNKWMLFFKLHGTH 662
Query 70 YVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGSVGGSTNVEKDEEQKEQ 129
++ GGK+ +I ++ + + NVK+ F +G S+ +K E ++
Sbjct 663 VAYEIYLGGKIIIKININKEEYNKMKENNINVKTFFNIYF--HKMGLSSAFQK--EAQKI 718
Query 130 LKHFDYQTETLIMGGRVPRDVSDPDSLAQWSDSVEELPMPVKLTLQPLEYLL 181
L F I+GG +V++ +W S+ MP++ L P + +
Sbjct 719 LNKFRISKHIAILGGNPGLNVNNTSFFEKWVHSINTNSMPIRTKLLPFSFFM 770
> tpv:TP03_0801 hypothetical protein
Length=353
Score = 58.9 bits (141), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 48/171 (28%), Positives = 74/171 (43%), Gaps = 10/171 (5%)
Query 14 TLAFNEAVAGLPPTFDGADEGKGCSPEVWRENTKDKLCADSTVKTWMNFIDQFGTHYVVR 73
T +F A++ L T D K C+ + N +K C + +K WM DQFGTH+
Sbjct 165 TRSFATAMSKL--TRDFKKHTKDCNAIKYSINKNNKDCKE--IKNWMELFDQFGTHFSYN 220
Query 74 LFAGGKLTH--QITMSNSDIAALSSLGTNVKSAL-KATFGVGSVGGSTNVEKDEEQKEQL 130
+ GG++T Q S + S+ V K GVG G V ++ + +
Sbjct 221 IKLGGRITFITQEEGSKDERGNEKSVDVGVGGKFEKDNKGVGIEGNVKFVFGNKRGESKN 280
Query 131 KHFDYQTETLIMGGRVPRDVSDPDSLAQWSDSVEELPMPVKLTLQPLEYLL 181
F Y T I+GG D+S +W SV + PMP++ P+ +
Sbjct 281 LSFKY---TNILGGLPVSDISKESEYVKWIKSVYKYPMPIRTQFAPISKIF 328
> bbo:BBOV_II002020 18.m06160; mac/perforin domain containing
protein
Length=420
Score = 56.2 bits (134), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 45/168 (26%), Positives = 75/168 (44%), Gaps = 8/168 (4%)
Query 14 TLAFNEAVAGLPPTFDGADEGKGCSPEVWRENTKDKLCADSTVKTWMNFIDQFGTHYVVR 73
T +F EA+ L P D K CS ++ C +K W+ F + FGTHYV +
Sbjct 197 TSSFMEAMKSLEPLPDTVK--KMCSAHDMIDDMTKSECI--PLKKWIKFFEMFGTHYVHQ 252
Query 74 LFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGSVGGSTNVEKDEEQKEQLKHF 133
L GGKL + ++ S + AL + +V + + G S + ++ + ++L
Sbjct 253 LLLGGKLIQTLKINASKLQALKNDSIDVDLVVSSVLGSSSASLLADKVVNKYKLDELG-- 310
Query 134 DYQTETLIMGGRVPRDVSDPDSLAQWSDSVEELPMPVKLTLQPLEYLL 181
++GG +P A W +SV E PMP+ + L+ L+
Sbjct 311 --AKSITVIGGNMPNTPITDAEYAIWGNSVAENPMPIGIVGDSLKNLM 356
> dre:100007284 Perforin-1-like
Length=574
Score = 49.7 bits (117), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 37/138 (26%), Positives = 70/138 (50%), Gaps = 16/138 (11%)
Query 59 WMNFIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSL-GTNVKSAL----KATFGVGS 113
+ + I +GTHY + GG++ IT + AA+S L T +K L ++ V +
Sbjct 202 YRSLIYTYGTHYTTNVKLGGQIK-AITAIKTCQAAVSGLTDTAIKDCLDVEASGSYSVAT 260
Query 114 VGGSTNVEKDEEQK----EQLKHFDYQTETLIMGGRVPRD------VSDPDSLAQWSDSV 163
V + K++++K ++ + +T I+GG + + S P++L W +S+
Sbjct 261 VKAEAHFCKEQQKKMGTNQKFSSMFSERQTDIIGGNINGEHLLFSGSSHPNALNSWLESL 320
Query 164 EELPMPVKLTLQPLEYLL 181
+ LP V+ +L+PL +LL
Sbjct 321 KSLPDVVQYSLKPLHFLL 338
> tpv:TP03_0810 hypothetical protein
Length=348
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 38/167 (22%), Positives = 72/167 (43%), Gaps = 19/167 (11%)
Query 17 FNEAVAGLPPTFDGADEGKGCSPEVWRENTKDKLCADSTVKTWMNFIDQFGTHYVVRLFA 76
F E V LP T + + C +++ T+ + C + ++ W+N +GTH
Sbjct 174 FVEMVGKLPNTVESTE----CPIDIFI--TELESCKN--LRIWINLFKTYGTHITTYAMT 225
Query 77 GGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGSVGGSTNVEKDEEQKEQLKHFDYQ 136
GGK + ++ N ++ A S N K+ K T G + E E + ++K +
Sbjct 226 GGKFINMESVVNINLQARDSDIKNTKA--KRT-------GEASSEFSEIFRRRIKGNKIK 276
Query 137 TETLIMGGRVPRDVS--DPDSLAQWSDSVEELPMPVKLTLQPLEYLL 181
++GG ++ DP ++W ++++ PMP+K L
Sbjct 277 KHLWVIGGSFVNNLEQIDPKMFSKWVKTIDKRPMPIKARFSELSMFF 323
> bbo:BBOV_III000320 17.m10445; hypothetical protein
Length=512
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 43/177 (24%), Positives = 76/177 (42%), Gaps = 25/177 (14%)
Query 9 FNWNYTLAFNEAVAGLPPTFDGADEGKG-CSPEVWRENTKDKLCADSTVKTWMNFIDQFG 67
F + F + V LP +E G C+P+V+ + C S++ W+ F ++G
Sbjct 125 FTGKISSVFIKEVEALPSI----NEDYGPCTPDVFIVDPLKSDC--SSMHQWVKFFKRYG 178
Query 68 THYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGSVGGSTNVEKDEEQK 127
TH + GG++ M + K T V ++GG ++ K ++
Sbjct 179 THMTSHITIGGQIISIDRMVEGE---------------KITKMVKTLGGGESIYKVTDKF 223
Query 128 EQLKHFDYQT-ETLIMGGRVPRDVSDPDSLA--QWSDSVEELPMPVKLTLQPLEYLL 181
+ ++ + LI+GG + DS A +WS SV E PMP++ LE+ +
Sbjct 224 SSVMTGKRESHQMLILGGYYFSGLESNDSKAFNRWSKSVWERPMPIRANFTSLEHFM 280
> dre:569443 Perforin-1-like
Length=588
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 36/146 (24%), Positives = 67/146 (45%), Gaps = 19/146 (13%)
Query 53 DSTVKTWMNFIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTN-VKSALKATFGV 111
+ T +++++ +++FGTHY++++ GG + +T +A L L + VK+ L
Sbjct 203 NQTKESYLSLVEKFGTHYIIQVKLGGGV-QSVTSVKQCMATLQDLSVDEVKTCLDVE-AS 260
Query 112 GSVGGSTNVEK-----DEEQKEQLKHFDYQTE-----TLIMGGRVPRDV------SDPDS 155
SV G +V+ + Q ++L + T I GG +DP +
Sbjct 261 ASVMGRISVDTAYRHCKQSQDKKLSAHSFANSFSDRLTEITGGHTQDSALLFSASNDPGA 320
Query 156 LAQWSDSVEELPMPVKLTLQPLEYLL 181
QW SV + P + +L+PL L+
Sbjct 321 YKQWLSSVPQQPDVISFSLKPLHMLM 346
> dre:100007189 Perforin-1-like
Length=574
Score = 42.7 bits (99), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 38/143 (26%), Positives = 67/143 (46%), Gaps = 26/143 (18%)
Query 59 WMNFIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGSVG--G 116
+ + I +GTHY + GG + + + +S+G +A+K V + G
Sbjct 202 YRSLISTYGTHYTTSVMLGGHMKAITAIKTCE----ASIGGLSDTAIKDCLDVEASGIYK 257
Query 117 STNVEKD----EEQKEQL--------KHFDYQTETLIMGGRVPRD------VSDPDSLAQ 158
+ VE + E K++L K + QTE I+GG + + S P++L
Sbjct 258 ALTVEAEARLCRELKQKLGTNLKMSSKFSERQTE--IIGGNINGEDLLFSGSSHPNALKG 315
Query 159 WSDSVEELPMPVKLTLQPLEYLL 181
W +S++ +P V+ TL+PL +LL
Sbjct 316 WLESLKSVPDVVQYTLKPLHFLL 338
> pfa:PF08_0052 perforin like protein 5
Length=676
Score = 41.6 bits (96), Expect = 0.001, Method: Composition-based stats.
Identities = 16/62 (25%), Positives = 35/62 (56%), Gaps = 2/62 (3%)
Query 32 DEGKGCSPEVWRENTKDKLCAD--STVKTWMNFIDQFGTHYVVRLFAGGKLTHQITMSNS 89
D+ CS + ++ N +K + T+ W++F + +GTH + ++ GGK+ H + N+
Sbjct 202 DDTYKCSLQYYKMNNMNKYSENCLKTITPWISFFNMYGTHVISGVYYGGKIIHNLYFENN 261
Query 90 DI 91
++
Sbjct 262 NL 263
> dre:100007241 Perforin-1-like
Length=1012
Score = 40.0 bits (92), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 37/143 (25%), Positives = 67/143 (46%), Gaps = 26/143 (18%)
Query 59 WMNFIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGSVG--G 116
+ + I +GTHY + GG + + + +S+G +A+K V + G
Sbjct 640 YRSLISTYGTHYTTSVMLGGHMKAITAIKTCE----ASIGGLSDTAIKDCLDVEASGIYK 695
Query 117 STNVEKD----EEQKEQL--------KHFDYQTETLIMGGRVPRD------VSDPDSLAQ 158
+ VE + E K++L K + QTE I+GG++ + S ++L
Sbjct 696 ALTVEAEARLCRELKQKLGTNLKMSSKFSERQTE--IIGGKINGEDLLFSGSSHKNALKG 753
Query 159 WSDSVEELPMPVKLTLQPLEYLL 181
W +S++ +P V+ TL+PL +LL
Sbjct 754 WLESLKSVPDVVQYTLKPLHFLL 776
> dre:100000903 Perforin-1-like; K07818 perforin 1
Length=580
Score = 39.7 bits (91), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 38/145 (26%), Positives = 60/145 (41%), Gaps = 29/145 (20%)
Query 59 WMNFIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTN-VKSAL----------KA 107
+ FI+ FGTHY+ ++ GG + H +T AL L T+ VK+ L KA
Sbjct 203 FYKFINTFGTHYITKVTLGGNV-HSVTSIRQCETALQGLSTDEVKACLDLEANMSILGKA 261
Query 108 TFGVGSVGGSTNVEKDEEQKEQLKHFD-----------YQTETLIMGGRVPRDVSDPDSL 156
S S N +K E + F+ + E L GG+ DP +
Sbjct 262 DMKAESKHCSQNQDKTESKTSFSSRFNDRLTEVTGGHTTEPELLFSGGK------DPSAY 315
Query 157 AQWSDSVEELPMPVKLTLQPLEYLL 181
+W S+ + P + +L+ L L+
Sbjct 316 KEWLASLPQNPDVISHSLESLHELI 340
> tpv:TP03_0799 hypothetical protein
Length=356
Score = 39.3 bits (90), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 38/175 (21%), Positives = 69/175 (39%), Gaps = 8/175 (4%)
Query 9 FNWNYTLAFNEAVAGLPPTFDGADEGKGCSPEVWRENTKDKLCADSTVKTWMNFIDQFGT 68
N+N L N + L + + + C ++R + D C + WM F +GT
Sbjct 142 LNFNGMLILN-VLKKLNKNCNSEFDNQKCPISMFRNDPFDANCI-RCIMPWMEFFKDYGT 199
Query 69 HYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVG-SVGGSTNVEKD-EEQ 126
+ GG + + + + F + S S N +K E
Sbjct 200 FMTKEITMGGVINKFYNIKKYEGSMRKEYKKKTIKQSSTFFHLSKSRSESLNEKKSGETN 259
Query 127 KEQLKHFDYQTETLIMGGRVPRDVSDPDSLAQWSDSVEELPMPVKLTLQPLEYLL 181
KE+L+ + TL +G P +VS+ ++ W + V P P+ L L P++ ++
Sbjct 260 KEELE----ELYTLTIGPEPPGNVSNSKVISDWLEKVVHNPTPIDLELVPIKQII 310
> hsa:732 C8B, MGC163447; complement component 8, beta polypeptide;
K03998 complement component 8 subunit beta
Length=591
Score = 37.4 bits (85), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 32/145 (22%), Positives = 58/145 (40%), Gaps = 25/145 (17%)
Query 59 WMNFIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVG------ 112
+ + FGTHY+ GG + + M+ + NV + K F +G
Sbjct 333 YRDLFRDFGTHYITEAVLGGIYEYTLVMNKEAMERGDYTLNNVHACAKNDFKIGGAIEEV 392
Query 113 ------SVG---GSTNVEKDEEQKEQLKHFDYQTETLIMGGR-------VPRDVSDPDSL 156
SVG G N KD +++ + L+ GG +++ D +
Sbjct 393 YVSLGVSVGKCRGILNEIKDRNKRDTMVE---DLVVLVRGGASEHITTLAYQELPTADLM 449
Query 157 AQWSDSVEELPMPVKLTLQPLEYLL 181
+W D+V+ P +K+ ++PL L+
Sbjct 450 QEWGDAVQYNPAIIKVKVEPLYELV 474
> mmu:18646 Prf1, Pfn, Pfp, Prf-1; perforin 1 (pore forming protein);
K07818 perforin 1
Length=554
Score = 37.4 bits (85), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 35/147 (23%), Positives = 61/147 (41%), Gaps = 22/147 (14%)
Query 54 STVKTWMNFIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGS 113
ST + I +GTH++ + GG+++ + + V L V S
Sbjct 206 STEHAYHRLISSYGTHFITAVDLGGRISVLTALRTCQLTLNGLTADEVGDCLNVEAQV-S 264
Query 114 VGGSTNVEKD----EEQKEQLK-----HFDYQTETL-IMGGRVPRDVSD---------PD 154
+G +V + EE+K+Q K H Y+ + ++GG P D + P+
Sbjct 265 IGAQASVSSEYKACEEKKKQHKMATSFHQTYRERHVEVLGG--PLDSTHDLLFGNQATPE 322
Query 155 SLAQWSDSVEELPMPVKLTLQPLEYLL 181
+ W+ S+ P V +L+PL LL
Sbjct 323 QFSTWTASLPSNPGLVDYSLEPLHTLL 349
> dre:559849 similar to perforin 1 (pore forming protein)
Length=555
Score = 37.0 bits (84), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 36/146 (24%), Positives = 62/146 (42%), Gaps = 20/146 (13%)
Query 53 DSTVKTWMN-FIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKA---- 107
DST ++ N FI +GTH++ R+ GG++ + ++ +V + L A
Sbjct 193 DSTSESAYNHFISIYGTHFLRRVDLGGRVKSTTAVRTCQVSMKGLSANDVSNCLSAEASA 252
Query 108 -TFGVGSVGGSTNVEKDEEQKEQLKHFDYQTE---TLIMGGR--------VPRDVSDPDS 155
GV G + + ++Q E+ F T I+GG P +VS
Sbjct 253 IIKGVKVSGQTAYCKSKQKQLERANSFSATFSDRVTDILGGNGEQQDILFTPSNVS---G 309
Query 156 LAQWSDSVEELPMPVKLTLQPLEYLL 181
+W S++++P V TL L L+
Sbjct 310 YGKWLGSLKKIPGVVSYTLSSLHMLV 335
> mmu:110382 C8b, 4930439B20Rik, AI595927; complement component
8, beta polypeptide; K03998 complement component 8 subunit
beta
Length=589
Score = 37.0 bits (84), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 25/142 (17%), Positives = 58/142 (40%), Gaps = 19/142 (13%)
Query 59 WMNFIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVG------ 112
+ + + FGTH++ GG + + M+ + ++V + +FG+G
Sbjct 332 YRDLLRDFGTHFITEAVLGGIYEYTLIMNKDAMEQGDYTLSHVTACAGGSFGIGGMVYKV 391
Query 113 ------SVGGSTNVEKDEEQKEQLKHFDYQTETLIMGGR-------VPRDVSDPDSLAQW 159
S +++ K+ ++ + L+ GG +++ P+ + W
Sbjct 392 YVKVGVSAKKCSDIMKEINERNKRSTMVEDLVVLVRGGTSEDITALAYKELPTPELMEAW 451
Query 160 SDSVEELPMPVKLTLQPLEYLL 181
D+V+ P +K+ +PL L+
Sbjct 452 GDAVKYNPAIIKIKAEPLYELV 473
> dre:795573 MGC163021; zgc:163021
Length=516
Score = 36.6 bits (83), Expect = 0.044, Method: Compositional matrix adjust.
Identities = 34/143 (23%), Positives = 64/143 (44%), Gaps = 15/143 (10%)
Query 53 DSTVKTWMNFIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLG-TNVKSALKATF-- 109
D T + I+ +GTHY+ ++ GG++ +T + +A L ++K L F
Sbjct 202 DQTKHLYQKMIETYGTHYIRQVHLGGRV-RWVTAFRTCLATLKGFSEIDIKHCLNMDFMI 260
Query 110 --GVGSVGGSTNVEKDEEQKEQLKHFDYQT----ETLIMGGR--VPRDV---SDPDSLAQ 158
G S + + + E++ YQT +T ++GG P V S ++ +Q
Sbjct 261 KLGFQQATASFSNKCSKILVEEMGMGFYQTFMTHKTEVLGGEKYFPELVLNQSPAEAYSQ 320
Query 159 WSDSVEELPMPVKLTLQPLEYLL 181
W S+ + P + + PL +L+
Sbjct 321 WRKSLHDNPDVISYAIFPLHHLV 343
> dre:100331881 hypothetical protein LOC100331881
Length=847
Score = 35.4 bits (80), Expect = 0.097, Method: Composition-based stats.
Identities = 34/135 (25%), Positives = 58/135 (42%), Gaps = 15/135 (11%)
Query 62 FIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTN-------VKSAL----KATFG 110
FI ++GTHY+ GG+ + + + + +S T+ VK L K T
Sbjct 303 FIQRYGTHYMEEGSLGGQYRALLELDANYMMEMSRTETDFHQCITRVKRRLFYKKKTTKC 362
Query 111 VGSVGGSTNVEKDEEQKEQLKHFDYQTETLIMGGRVPRDVSDPDSLAQ----WSDSVEEL 166
V + N ++ K +K + + G D+ +PD+ Q W+ SV+E
Sbjct 363 VKLMKTIENFSENRNHKMPIKTDIIGGNSAYIAGLSLLDLENPDNNKQMYTKWAGSVKEF 422
Query 167 PMPVKLTLQPLEYLL 181
P +K L+PL L+
Sbjct 423 PKVIKQKLRPLHELV 437
> dre:570832 c7, cb349, wu:fa01f03; complement component 7; K03996
complement component 7
Length=820
Score = 35.4 bits (80), Expect = 0.10, Method: Composition-based stats.
Identities = 34/135 (25%), Positives = 58/135 (42%), Gaps = 15/135 (11%)
Query 62 FIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTN-------VKSAL----KATFG 110
FI ++GTHY+ GG+ + + + + +S T+ VK L K T
Sbjct 276 FIQRYGTHYMEEGSLGGQYRALLELDANYMMEMSRTETDFHQCITRVKRRLFYKKKTTKC 335
Query 111 VGSVGGSTNVEKDEEQKEQLKHFDYQTETLIMGGRVPRDVSDPDSLAQ----WSDSVEEL 166
V + N ++ K +K + + G D+ +PD+ Q W+ SV+E
Sbjct 336 VKLMKTIENFSENRNHKMPIKTDIIGGNSAYIAGLSLLDLENPDNNKQMYTKWAGSVKEF 395
Query 167 PMPVKLTLQPLEYLL 181
P +K L+PL L+
Sbjct 396 PKVIKQKLRPLHELV 410
> dre:100332024 hypothetical protein LOC100332024
Length=820
Score = 35.4 bits (80), Expect = 0.10, Method: Composition-based stats.
Identities = 34/135 (25%), Positives = 58/135 (42%), Gaps = 15/135 (11%)
Query 62 FIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTN-------VKSAL----KATFG 110
FI ++GTHY+ GG+ + + + + +S T+ VK L K T
Sbjct 276 FIQRYGTHYMEEGSLGGQYRALLELDANYMMEMSRTETDFHQCITRVKRRLFYKKKTTKC 335
Query 111 VGSVGGSTNVEKDEEQKEQLKHFDYQTETLIMGGRVPRDVSDPDSLAQ----WSDSVEEL 166
V + N ++ K +K + + G D+ +PD+ Q W+ SV+E
Sbjct 336 VKLMKTIENFSENRNHKMPIKTDIIGGNSAYIAGLSLLDLENPDNNKQMYTKWAGSVKEF 395
Query 167 PMPVKLTLQPLEYLL 181
P +K L+PL L+
Sbjct 396 PKVIKQKLRPLHELV 410
> dre:559384 si:ch211-103n10.4
Length=517
Score = 33.1 bits (74), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 33/133 (24%), Positives = 62/133 (46%), Gaps = 15/133 (11%)
Query 63 IDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLG-TNVKSA----LKATFGVGSVGGS 117
I+ +GTHY+ ++ GG++ ++T + +A L T++K+ LK T G S
Sbjct 213 IETYGTHYIRQVHLGGRV-RRVTAFRTCLATLKGFSETDIKNCLNVELKITLGFLPGNAS 271
Query 118 TNVEKDEEQKEQLKHFDYQ----TETLIMGGR--VPRDV---SDPDSLAQWSDSVEELPM 168
+ + + K+ L YQ +T ++GG P V S ++ + W S+ P
Sbjct 272 LSNKCSQILKDNLSMGFYQGFMTHKTEVLGGEKYFPDLVLSQSPAEAYSNWMMSLHNNPD 331
Query 169 PVKLTLQPLEYLL 181
+ + PL +L+
Sbjct 332 VISYAIFPLHHLV 344
> dre:445102 c8a, zgc:92465; complement component 8, alpha polypeptide;
K03997 complement component 8 subunit alpha
Length=478
Score = 32.0 bits (71), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 30/142 (21%), Positives = 55/142 (38%), Gaps = 24/142 (16%)
Query 59 WMNFIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGV------- 111
+ F +++GTHYV GG + + ++ +++ G + S + +FG+
Sbjct 226 YSQFFNEYGTHYVTEGTMGGLMDYVAVVNINEMEENQMTGQMIGSCIGGSFGLVFMEKIK 285
Query 112 -----GSVGGSTNVEK--DEEQKEQLKHFDYQTETLIMGGRVPRD-----VSDPDSLAQW 159
S G T+ EK DE F + + GG + D S W
Sbjct 286 ATVKGKSCGKFTSNEKTSDESHSAIKDVFGF-----VKGGNTASSAGSLGIKDAKSYKDW 340
Query 160 SDSVEELPMPVKLTLQPLEYLL 181
S++ P ++ + P+ LL
Sbjct 341 GKSLKYNPALIEFEILPIYELL 362
> xla:443975 c8a, MGC80388; complement component 8, alpha polypeptide;
K03997 complement component 8 subunit alpha
Length=589
Score = 32.0 bits (71), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 20/93 (21%), Positives = 38/93 (40%), Gaps = 1/93 (1%)
Query 59 WMNFIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGSVGGST 118
+ FI+ FGTHY+ GG + + + + ++ + V A+ G+ +
Sbjct 332 YAQFINDFGTHYITSGIMGGVMENVVVLDKEEMKRQEITASMVSHCFGASIGLSVNSDQS 391
Query 119 NVEKDEE-QKEQLKHFDYQTETLIMGGRVPRDV 150
++ + E K F+ Q E R +DV
Sbjct 392 DLLPSAKLSGEICKKFEKQNEDNSSSSRTIKDV 424
> xla:432189 c7, MGC82368; complement component 7; K03996 complement
component 7
Length=830
Score = 31.6 bits (70), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 34/141 (24%), Positives = 61/141 (43%), Gaps = 19/141 (13%)
Query 58 TWMNFIDQFGTHYVVRLFAGGKL-------THQIT---MSNSDIAALSSLGTN---VKSA 104
T+ FID++GTH++ GG+ + +IT ++ D+ + +S N VK +
Sbjct 276 TYRKFIDKYGTHFLQSGSLGGEYKFLFYLDSEKITANGVTKRDMQSCTSSSKNFFFVKYS 335
Query 105 LKATFGVGSVGGSTNVEKDEEQKEQLKHFDYQTETLIMGG----RVPRDVSDPDSLAQWS 160
K + V ++ D E + + F E + G + +S+ + A W+
Sbjct 336 SKECKALNEVIRQSSGSTDREVRGDV--FVQGGEAKFVAGLSYFSLDNPLSNENRYAAWA 393
Query 161 DSVEELPMPVKLTLQPLEYLL 181
SV LP +K PL L+
Sbjct 394 GSVSNLPSVIKHKTTPLYELV 414
> ath:AT1G29690 CAD1; CAD1 (constitutively activated cell death
1)
Length=561
Score = 31.6 bits (70), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 30/121 (24%), Positives = 51/121 (42%), Gaps = 12/121 (9%)
Query 61 NFIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGSVGGSTNV 120
+FI+ +GTH V + GG+ I S +S + V +K F +
Sbjct 176 SFIENYGTHIVTSVTIGGRDVVYIRQHQSSPLPVSEIENYVNDMIKHRF---------HE 226
Query 121 EKDEEQKEQLKHFDYQTETLIMGGRVPRDVSDPDSLAQWSDSVEELPMPVKLTLQPLEYL 180
+ + LK+ D T+I R D+ S A+W+++V P + +T P+ L
Sbjct 227 AESQSITGPLKYKDKDI-TVIFRRRGGDDLE--QSHARWAETVPAAPDIINMTFTPIVSL 283
Query 181 L 181
L
Sbjct 284 L 284
> tpv:TP02_0641 hypothetical protein
Length=125
Score = 31.6 bits (70), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 9/29 (31%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 53 DSTVKTWMNFIDQFGTHYVVRLFAGGKLT 81
+ +K + F + +GTHY++++ GGK+
Sbjct 3 ECVIKEYKKFFELYGTHYIIKIVLGGKIV 31
> mmu:11832 Aqp7, AQP7L, AQPap; aquaporin 7; K08771 aquaporin-7
Length=303
Score = 31.2 bits (69), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 18/62 (29%), Positives = 28/62 (45%), Gaps = 4/62 (6%)
Query 62 FIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGSVGGSTNVE 121
F+ +F + YV+ +F G + H + NS S LG N+ T GV GG +
Sbjct 22 FLAEFLSTYVMMVFGLGSVAHMVLGENSG----SYLGVNLGFGFGVTMGVHVAGGISGAH 77
Query 122 KD 123
+
Sbjct 78 MN 79
> xla:379504 c9, MGC64276; complement component 9; K04000 complement
component 9
Length=593
Score = 30.8 bits (68), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 30/153 (19%), Positives = 60/153 (39%), Gaps = 30/153 (19%)
Query 59 WMNFIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALK----ATFGVGSV 114
+ + ++ +GTHY V GGK + + ++ + ++K L+ A GV +
Sbjct 319 YFSILEMYGTHYSVSGNLGGKYELVYVLDSIEMNSRELTTEDIKDCLRFNADAGIGVKAE 378
Query 115 GGSTNV--------------EKDEEQKEQLKHFDYQTETLIMGGRV------------PR 148
G + ++ E + E +K + + GG V +
Sbjct 379 GANLDLNPKIKGDVCKTGGGESETEPSINIKPVIESIISFVDGGTVEYVTALEEKLNKKQ 438
Query 149 DVSDPDSLAQWSDSVEELPMPVKLTLQPLEYLL 181
V+D + QW+ S++E P +K P+ L+
Sbjct 439 PVADVNDYVQWASSLKEAPAVIKSKPNPIYSLI 471
> pfa:PF08_0050 MAC/Perforin, putative
Length=654
Score = 30.8 bits (68), Expect = 2.4, Method: Composition-based stats.
Identities = 14/47 (29%), Positives = 25/47 (53%), Gaps = 3/47 (6%)
Query 32 DEGKGCSPEVWRENTKDKLCADSTVKTWMNFIDQFGTHYVVRLFAGG 78
++ K C+ ++ N L +K+WM F +++GTH V+ GG
Sbjct 181 NQMKLCTKVLYMNNN---LHCSEGIKSWMKFFEKYGTHVVLSAHFGG 224
> dre:560278 hypothetical LOC560278
Length=995
Score = 30.8 bits (68), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 31/122 (25%), Positives = 53/122 (43%), Gaps = 10/122 (8%)
Query 66 FGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGSVGGSTNVEKDEE 125
GTH V + G S S++ VK+AL+ + SVG + +E
Sbjct 142 IGTHVVTAILYGANACFVFDRDVSKEEDESNIKGEVKAALEKLQSIVSVGAKAKITSNEN 201
Query 126 QKEQLKHFDYQTETLIMGGRVPRD-VSDPDSLAQWSD------SVEELPMPVKLTLQPLE 178
QK+ + F T T ++P + + D+L ++D +EL +P+++ L PL
Sbjct 202 QKDAVNKF---TCTFHGDFQLPFNPTTFEDALQVFADLPKLLKESQELAVPLRVWLYPLN 258
Query 179 YL 180
L
Sbjct 259 KL 260
> cel:T18D3.4 myo-2; MYOsin heavy chain structural genes family
member (myo-2); K10352 myosin heavy chain
Length=1947
Score = 30.4 bits (67), Expect = 3.1, Method: Composition-based stats.
Identities = 22/90 (24%), Positives = 44/90 (48%), Gaps = 3/90 (3%)
Query 53 DSTVKTWMNFIDQFGTHYVVRLFAGGKLTHQITMSNSDIA-ALSSLGTNVKSALKATFGV 111
+S+VK ++ + V A +L ++I + NSDIA A + L + +A
Sbjct 1719 ESSVKEHQEHNNELNSQNVALAAAKSQLDNEIALLNSDIAEAHTELSASEDRGRRAASDA 1778
Query 112 GSVGGSTNVEKDEEQKEQLKHFDYQTETLI 141
+ + ++ ++EQ +QL+ F Q E+ +
Sbjct 1779 AKL--AEDLRHEQEQSQQLERFKKQLESAV 1806
> mmu:214642 A430107O13Rik, 6720481P07, AI552584; RIKEN cDNA A430107O13
gene
Length=1026
Score = 30.4 bits (67), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 24/50 (48%), Gaps = 2/50 (4%)
Query 4 AQTDNFNWNYTLAFNEAVAGLPPTFDGADEGKGCSPEVWRENTKDKLCAD 53
AQ WN + NE + P + +G GK +P V EN KD C+D
Sbjct 515 AQLQPLEWN-SPTRNETIEEPPRSLEGIQSGKDAAPRVLFEN-KDIHCSD 562
> cpv:cgd4_860 possible WWE domain
Length=1246
Score = 30.4 bits (67), Expect = 3.7, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 27/57 (47%), Gaps = 1/57 (1%)
Query 3 LAQTDNFN-WNYTLAFNEAVAGLPPTFDGADEGKGCSPEVWRENTKDKLCADSTVKT 58
L+ +N++ N TL + A PP D + P + EN K L +DST+ T
Sbjct 210 LSSNNNYHRCNTTLQIDRTFAAKPPNGGNLDRKQLIIPRLQIENCKGNLLSDSTINT 266
> ath:AT4G24290 hypothetical protein
Length=350
Score = 29.6 bits (65), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 38/162 (23%), Positives = 59/162 (36%), Gaps = 22/162 (13%)
Query 38 SPEVWRENTKDKLCADSTVKTWMNFIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSL 97
S + RE+ K + + FID +GTH +V + GGK +S L
Sbjct 145 SQVLLREHVKQAVPSTWDPAALARFIDIYGTHIIVSVKMGGKDVIYAKQQHSSKLQPEDL 204
Query 98 GTNVKSALKATFGVGSVGGSTNVEKDE-----EQKEQLKHF-------------DYQTET 139
+K F SV +T E+ + E KEQ F DY
Sbjct 205 QKRLKEVADKRFVEASVVHNTGSERVQASSKVETKEQRLRFADTSSLGSYANKEDYVFMC 264
Query 140 LIMGGRVPRDVSDPDSLAQWSDSVEELPMPVKLTLQPLEYLL 181
GG R++ +W +V+ P + ++ P+ LL
Sbjct 265 KRRGGNDNRNLMH----NEWLQTVQMEPDVISMSFIPITSLL 302
> hsa:5551 PRF1, FLH2, HPLH2, MGC65093, P1, PFN1, PFP; perforin
1 (pore forming protein); K07818 perforin 1
Length=555
Score = 29.6 bits (65), Expect = 6.3, Method: Composition-based stats.
Identities = 31/145 (21%), Positives = 65/145 (44%), Gaps = 18/145 (12%)
Query 54 STVKTWMNFIDQFGTHYVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSAL----KATF 109
ST ++ I +GTH++ + GG+++ + ++A V+ L +
Sbjct 207 STQPAYLRLISNYGTHFIRAVELGGRISALTALRTCELALEGLTDNEVEDCLTVEAQVNI 266
Query 110 GV-GSVGGSTNVEKDEEQKEQLKHFDYQT----ETLIMGGRVPRDVSD--------PDSL 156
G+ GS+ +++++K ++ +QT + ++GG ++D P+
Sbjct 267 GIHGSISAEAKACEEKKKKHKMTASFHQTYRERHSEVVGGH-HTSINDLLFGIQAGPEQY 325
Query 157 AQWSDSVEELPMPVKLTLQPLEYLL 181
+ W +S+ P V TL+PL LL
Sbjct 326 SAWVNSLPGSPGLVDYTLEPLHVLL 350
> pfa:PFF0935c conserved Plasmodium protein, unknown function
Length=3618
Score = 29.3 bits (64), Expect = 7.2, Method: Composition-based stats.
Identities = 17/82 (20%), Positives = 42/82 (51%), Gaps = 3/82 (3%)
Query 63 IDQFGTH---YVVRLFAGGKLTHQITMSNSDIAALSSLGTNVKSALKATFGVGSVGGSTN 119
I+Q G H + + + G+++H+ N +I+ + + N + + + + + N
Sbjct 933 INQKGNHNEEILHKEISHGEISHKEISHNDEISPIDKIYHNDEISYHDGVNMNTCEQNNN 992
Query 120 VEKDEEQKEQLKHFDYQTETLI 141
++ E+QKEQ++ +D + +I
Sbjct 993 IKLREKQKEQIEKYDQTIKYII 1014
> dre:100007639 pancreatic secretory granule membrane major glycoprotein
GP2-like
Length=1301
Score = 29.3 bits (64), Expect = 7.3, Method: Composition-based stats.
Identities = 19/65 (29%), Positives = 30/65 (46%), Gaps = 2/65 (3%)
Query 116 GSTNVEKDEEQKEQLK--HFDYQTETLIMGGRVPRDVSDPDSLAQWSDSVEELPMPVKLT 173
GS + E D+E +++ D + L+M V+DPDS +W E P P T
Sbjct 1125 GSVDAELDQEMHVEVRVEGVDSRQFALVMDTCWATPVNDPDSSLRWDLIAHECPNPADET 1184
Query 174 LQPLE 178
+ L+
Sbjct 1185 VDLLQ 1189
> tpv:TP03_0775 small GTP-binding protein Rab1; K07976 Rab family,
other
Length=227
Score = 29.3 bits (64), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 12/33 (36%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 48 DKLCADSTVKTWMNFIDQFGTHYVVRLFAGGKL 80
+KL D +TW+ ID++ T V +L G K+
Sbjct 98 NKLSFDHITETWLQDIDKYATSNVCKLLIGNKI 130
Lambda K H
0.314 0.131 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4924747408
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40