bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0011_orf2
Length=266
Score E
Sequences producing significant alignments: (Bits) Value
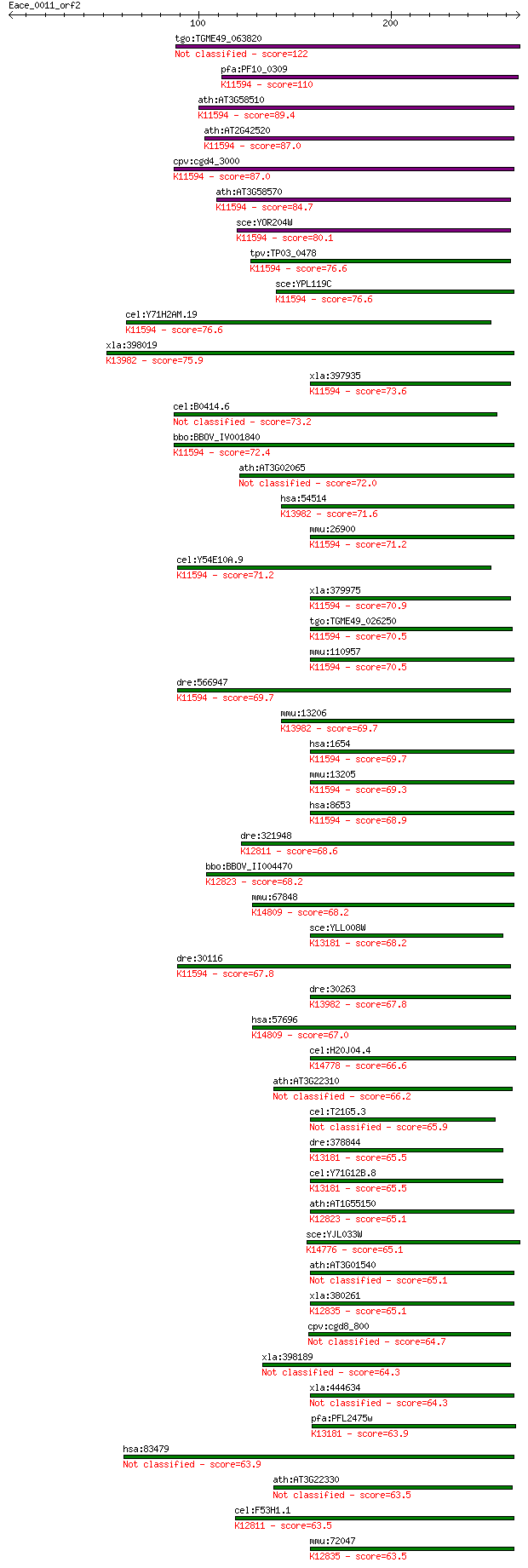
tgo:TGME49_063820 ATP-dependent RNA helicase, putative (EC:2.7... 122 9e-28
pfa:PF10_0309 DEAD/DEAH box helicase, putative; K11594 ATP-dep... 110 7e-24
ath:AT3G58510 DEAD box RNA helicase, putative (RH11); K11594 A... 89.4 1e-17
ath:AT2G42520 DEAD box RNA helicase, putative; K11594 ATP-depe... 87.0 7e-17
cpv:cgd4_3000 Dbp1p, eIF4a-1 family RNA SFII helicase (DEXDC+H... 87.0 7e-17
ath:AT3G58570 DEAD box RNA helicase, putative; K11594 ATP-depe... 84.7 3e-16
sce:YOR204W DED1, SPP81; ATP-dependent DEAD (Asp-Glu-Ala-Asp)-... 80.1 7e-15
tpv:TP03_0478 RNA helicase; K11594 ATP-dependent RNA helicase ... 76.6 8e-14
sce:YPL119C DBP1, LPH8; Dbp1p (EC:3.6.1.-); K11594 ATP-depende... 76.6 8e-14
cel:Y71H2AM.19 hypothetical protein; K11594 ATP-dependent RNA ... 76.6 9e-14
xla:398019 ddx4, vasa, vlg1; DEAD (Asp-Glu-Ala-Asp) box polype... 75.9 1e-13
xla:397935 an3; ATP dependent RNA helicase (EC:3.6.4.13); K115... 73.6 8e-13
cel:B0414.6 glh-3; Germ-Line Helicase family member (glh-3) 73.2 1e-12
bbo:BBOV_IV001840 21.m02846; DEAD/DEAH box helicase (EC:3.6.1.... 72.4 1e-12
ath:AT3G02065 DEAD/DEAH box helicase family protein 72.0 2e-12
hsa:54514 DDX4, MGC111074, VASA; DEAD (Asp-Glu-Ala-Asp) box po... 71.6 3e-12
mmu:26900 Ddx3y, 8030469F12Rik, D1Pas1-rs1, Dby; DEAD (Asp-Glu... 71.2 3e-12
cel:Y54E10A.9 vbh-1; Vasa- and Belle-like Helicase family memb... 71.2 3e-12
xla:379975 ddx3x, MGC52935, pl10; DEAD (Asp-Glu-Ala-Asp) box p... 70.9 4e-12
tgo:TGME49_026250 ATP-dependent RNA helicase, putative ; K1159... 70.5 6e-12
mmu:110957 D1Pas1, AU016353, Pl10; DNA segment, Chr 1, Pasteur... 70.5 7e-12
dre:566947 ddx3, fb74g09, wu:fb74g09, zgc:158804; DEAD (Asp-Gl... 69.7 1e-11
mmu:13206 Ddx4, AV206478, Mvh, VASA; DEAD (Asp-Glu-Ala-Asp) bo... 69.7 1e-11
hsa:1654 DDX3X, DBX, DDX14, DDX3, HLP2; DEAD (Asp-Glu-Ala-Asp)... 69.7 1e-11
mmu:13205 Ddx3x, D1Pas1-rs2, Ddx3, Fin14; DEAD/H (Asp-Glu-Ala-... 69.3 1e-11
hsa:8653 DDX3Y, DBY; DEAD (Asp-Glu-Ala-Asp) box polypeptide 3,... 68.9 2e-11
dre:321948 ddx46, fb39a03, wu:fb39a03; DEAD (Asp-Glu-Ala-Asp) ... 68.6 2e-11
bbo:BBOV_II004470 18.m06373; p68-like protein; K12823 ATP-depe... 68.2 3e-11
mmu:67848 Ddx55, 2810021H22Rik, MGC143677, MGC143678, mKIAA159... 68.2 3e-11
sce:YLL008W DRS1; Drs1p (EC:3.6.1.-); K13181 ATP-dependent RNA... 68.2 3e-11
dre:30116 pl10, etID309900.24, p110, p110a, pl10a, wu:fb43h11,... 67.8 4e-11
dre:30263 vasa, MGC158535, fi24g05, vas, vlg, wu:fi24g05, zgc:... 67.8 4e-11
hsa:57696 DDX55, FLJ16577, KIAA1595, MGC33209; DEAD (Asp-Glu-A... 67.0 6e-11
cel:H20J04.4 hypothetical protein; K14778 ATP-dependent RNA he... 66.6 8e-11
ath:AT3G22310 PMH1; PMH1 (PUTATIVE MITOCHONDRIAL RNA HELICASE ... 66.2 1e-10
cel:T21G5.3 glh-1; Germ-Line Helicase family member (glh-1) 65.9 2e-10
dre:378844 ddx27, cb843; DEAD (Asp-Glu-Ala-Asp) box polypeptid... 65.5 2e-10
cel:Y71G12B.8 hypothetical protein; K13181 ATP-dependent RNA h... 65.5 2e-10
ath:AT1G55150 DEAD box RNA helicase, putative (RH20); K12823 A... 65.1 2e-10
sce:YJL033W HCA4, DBP4, ECM24; Hca4p (EC:3.6.1.-); K14776 ATP-... 65.1 2e-10
ath:AT3G01540 DRH1; DRH1 (DEAD BOX RNA HELICASE 1); ATP-depend... 65.1 3e-10
xla:380261 ddx42, MGC54025; DEAD (Asp-Glu-Ala-Asp) box polypep... 65.1 3e-10
cpv:cgd8_800 Prp5p C terminal KH. eIF4A-1-family RNA SFII heli... 64.7 3e-10
xla:398189 RNA helicase II/Gu 64.3 4e-10
xla:444634 MGC84147 protein 64.3 4e-10
pfa:PFL2475w DEAD/DEAH box helicase, putative; K13181 ATP-depe... 63.9 5e-10
hsa:83479 DDX59, DKFZp564B1023, ZNHIT5; DEAD (Asp-Glu-Ala-Asp)... 63.9 6e-10
ath:AT3G22330 PMH2; PMH2 (putative mitochondrial RNA helicase ... 63.5 7e-10
cel:F53H1.1 hypothetical protein; K12811 ATP-dependent RNA hel... 63.5 8e-10
mmu:72047 Ddx42, 1810047H21Rik, AW319508, AW556242, B430002H05... 63.5 8e-10
> tgo:TGME49_063820 ATP-dependent RNA helicase, putative (EC:2.7.11.1)
Length=1053
Score = 122 bits (307), Expect = 9e-28, Method: Compositional matrix adjust.
Identities = 69/182 (37%), Positives = 103/182 (56%), Gaps = 4/182 (2%)
Query 88 NEDELFDLS-LTGGMFFDLLEQEAVCTCVGGDIEAPEPAETLEQCSE--GDSSLLAPRVV 144
N DE ++ G L +QE V G + +PA ++ G +L ++
Sbjct 601 NPDENYNADERLGDAIGQLEDQEVVIAFAGPTADVHDPASLPLPVADVVGSERVLHSAIL 660
Query 145 ENLKNNLKCRWLLPIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRP 204
NL L + L+P+Q+++I + G D+ A+TG+GKTLAFL P+ S+ L + RP
Sbjct 661 GNLLF-LGFKRLMPVQRHSIPIGCAGYDICGSASTGSGKTLAFLVPMASRLLKMSPINRP 719
Query 205 HFAGSHAQASPLAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREINQK 264
F G HAQASP ++L PTRELA+QI E + R+ GT L+H + IGG+ +QV IN++
Sbjct 720 FFPGPHAQASPTGLILVPTRELALQIGEDIVRVCRGTGLTHILLIGGMPQKDQVDHINRQ 779
Query 265 QI 266
QI
Sbjct 780 QI 781
> pfa:PF10_0309 DEAD/DEAH box helicase, putative; K11594 ATP-dependent
RNA helicase [EC:3.6.4.13]
Length=917
Score = 110 bits (274), Expect = 7e-24, Method: Composition-based stats.
Identities = 60/155 (38%), Positives = 90/155 (58%), Gaps = 6/155 (3%)
Query 112 CTCVGGD-IEAPEPAETLEQCSEGDSSLLAPRVVENLKNNLKCRWLLPIQKYAIAVAAKG 170
C G + I PEP +EQ S+ L + +N+ C + IQKY+I V K
Sbjct 509 CKLYGDEKINIPEPLINMEQLSK----LCDEELYKNICLKGYCN-MTTIQKYSIPVILKK 563
Query 171 LDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQASPLAVVLCPTRELAIQI 230
++L+AC+ TG+GKT +FL P++S ++ RPHF GS+A SPL ++LCPTREL IQI
Sbjct 564 INLLACSQTGSGKTFSFLCPIISNLKKENEILRPHFPGSYACISPLCLILCPTRELVIQI 623
Query 231 NEQLARLVVGTTLSHSIWIGGVSATEQVREINQKQ 265
++ L ++ + GG + +QV +IN+KQ
Sbjct 624 QNEVNSLTKNMSIVCMSFYGGETMKDQVAQINEKQ 658
> ath:AT3G58510 DEAD box RNA helicase, putative (RH11); K11594
ATP-dependent RNA helicase [EC:3.6.4.13]
Length=612
Score = 89.4 bits (220), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 59/166 (35%), Positives = 87/166 (52%), Gaps = 17/166 (10%)
Query 100 GMFFDLLEQEAVCTCVGGDIEAPEPAETLEQCSEGDSSLLAPRVVENLKNNLKCRWL--L 157
G+ FD E V T GGD+ P P T GD+ L N +C+++
Sbjct 128 GINFDAYEDIPVETS-GGDV--PPPVNTFADIDLGDALNL---------NIRRCKYVRPT 175
Query 158 PIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQASPLA 217
P+Q++AI + DL+ACA TG+GKT AF P++S + Q + RP GS A P A
Sbjct 176 PVQRHAIPILLAERDLMACAQTGSGKTAAFCFPIISGIMKDQHVERPR--GSRA-VYPFA 232
Query 218 VVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREINQ 263
V+L PTRELA QI+++ + T + + GG +Q+RE+ +
Sbjct 233 VILSPTRELACQIHDEAKKFSYQTGVKVVVAYGGTPIHQQLRELER 278
> ath:AT2G42520 DEAD box RNA helicase, putative; K11594 ATP-dependent
RNA helicase [EC:3.6.4.13]
Length=633
Score = 87.0 bits (214), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 56/163 (34%), Positives = 86/163 (52%), Gaps = 17/163 (10%)
Query 103 FDLLEQEAVCTCVGGDIEAPEPAETLEQCSEGDSSLLAPRVVENLKNNLKCRWL--LPIQ 160
FD E + T GD P P T + G++ L N +C+++ P+Q
Sbjct 139 FDAYEDIPIETS--GD-NVPPPVNTFAEIDLGEALNL---------NIRRCKYVKPTPVQ 186
Query 161 KYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQASPLAVVL 220
++AI + +G DL+ACA TG+GKT AF P++S + Q + RP GS PLAV+L
Sbjct 187 RHAIPILLEGRDLMACAQTGSGKTAAFCFPIISGIMKDQHVQRPR--GSRT-VYPLAVIL 243
Query 221 CPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREINQ 263
PTRELA QI+++ + T + + GG +Q+RE+ +
Sbjct 244 SPTRELASQIHDEAKKFSYQTGVKVVVAYGGTPINQQLRELER 286
> cpv:cgd4_3000 Dbp1p, eIF4a-1 family RNA SFII helicase (DEXDC+HELICc)
; K11594 ATP-dependent RNA helicase [EC:3.6.4.13]
Length=702
Score = 87.0 bits (214), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 59/181 (32%), Positives = 94/181 (51%), Gaps = 11/181 (6%)
Query 87 DNEDELFDLSLT--GGMFFDLLEQEAVCTCVGGDIEAPEPAETLEQCSEGDSSLLAPRVV 144
D+ED++F S G+ FD + V G D +P ++ + EG +L +
Sbjct 160 DDEDKIFSKSKEHRAGINFDAYDNIPV-EMTGSDTNKIKPMQSFMEL-EGIHEIL----L 213
Query 145 ENLKNNLKCRWLLPIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRP 204
+N++ +K P+QK++I G DL+ACA TG+GKT AFL P+V K L P
Sbjct 214 DNIRR-VKYERPTPVQKFSIPTVLNGRDLMACAQTGSGKTAAFLFPIVMKMLNDGPPPTP 272
Query 205 HFAGSHAQ--ASPLAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREIN 262
+ + A P+A+VL PTRELAIQ E+ + GT + ++ GG Q+ +++
Sbjct 273 QQSSLRIKRMAYPVALVLSPTRELAIQTYEESRKFCFGTGIRTNVLYGGSEVRSQIMDLD 332
Query 263 Q 263
+
Sbjct 333 R 333
> ath:AT3G58570 DEAD box RNA helicase, putative; K11594 ATP-dependent
RNA helicase [EC:3.6.4.13]
Length=646
Score = 84.7 bits (208), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 52/155 (33%), Positives = 79/155 (50%), Gaps = 15/155 (9%)
Query 109 EAVCTCVGGDIEAPEPAETLEQCSEGDSSLLAPRVVENLKNNLKCRWL--LPIQKYAIAV 166
E + GD P P T + G++ L N +C+++ P+Q+ AI +
Sbjct 130 EDIPIETSGD-NVPPPVNTFAEIDLGEALNL---------NIQRCKYVKPTPVQRNAIPI 179
Query 167 AAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQASPLAVVLCPTREL 226
A G DL+ACA TG+GKT AF P++S + Q + RP PLAV+L PTREL
Sbjct 180 LAAGRDLMACAQTGSGKTAAFCFPIISGIMKDQHIERPRGVRG---VYPLAVILSPTREL 236
Query 227 AIQINEQLARLVVGTTLSHSIWIGGVSATEQVREI 261
A QI+++ + T + + GG +Q+RE+
Sbjct 237 ACQIHDEARKFSYQTGVKVVVAYGGTPVNQQIREL 271
> sce:YOR204W DED1, SPP81; ATP-dependent DEAD (Asp-Glu-Ala-Asp)-box
RNA helicase, required for translation initiation of all
yeast mRNAs; mutations in human DEAD-box DBY are a frequent
cause of male infertility (EC:3.6.1.-); K11594 ATP-dependent
RNA helicase [EC:3.6.4.13]
Length=604
Score = 80.1 bits (196), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 52/146 (35%), Positives = 77/146 (52%), Gaps = 13/146 (8%)
Query 120 EAPEPAETLEQCSEGDSSLLAPRVVENLKNNLKCRWL--LPIQKYAIAVAAKGLDLVACA 177
+ PEP +E S L ++EN+K R+ P+QKY++ + A G DL+ACA
Sbjct 136 DVPEP------ITEFTSPPLDGLLLENIK---LARFTKPTPVQKYSVPIVANGRDLMACA 186
Query 178 ATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQ--ASPLAVVLCPTRELAIQINEQLA 235
TG+GKT FL P++S+ +P GS Q A P AV++ PTRELA QI ++
Sbjct 187 QTGSGKTGGFLFPVLSESFKTGPSPQPESQGSFYQRKAYPTAVIMAPTRELATQIFDEAK 246
Query 236 RLVVGTTLSHSIWIGGVSATEQVREI 261
+ + + + GG Q+REI
Sbjct 247 KFTYRSWVKACVVYGGSPIGNQLREI 272
> tpv:TP03_0478 RNA helicase; K11594 ATP-dependent RNA helicase
[EC:3.6.4.13]
Length=741
Score = 76.6 bits (187), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 49/139 (35%), Positives = 77/139 (55%), Gaps = 8/139 (5%)
Query 127 TLEQCSEGDSSLLAPRVVENLKNNLKCRWL--LPIQKYAIAVAAKGLDLVACAATGTGKT 184
+++ E DSS+ P+++ N++ K + PIQK++I V G DL+ACA TG+GKT
Sbjct 219 SIKPIEEFDSSV-HPKLIPNIR---KVNYTKPTPIQKHSIPVILAGRDLMACAQTGSGKT 274
Query 185 LAFLSPLVSKCLLHQQLFRPHFAGSHAQ--ASPLAVVLCPTRELAIQINEQLARLVVGTT 242
AFL P+V+ L +P + + A P+ +VL PTRELA+QI + + GT
Sbjct 275 AAFLLPIVTSMLRTGPPKQPTLSPLYGARVALPVCLVLSPTRELAVQIFSESRKFNFGTG 334
Query 243 LSHSIWIGGVSATEQVREI 261
+ + GG Q+ E+
Sbjct 335 IRTVVLYGGSEVRRQLIEL 353
> sce:YPL119C DBP1, LPH8; Dbp1p (EC:3.6.1.-); K11594 ATP-dependent
RNA helicase [EC:3.6.4.13]
Length=617
Score = 76.6 bits (187), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 50/135 (37%), Positives = 71/135 (52%), Gaps = 17/135 (12%)
Query 140 APRVVENLKNNLKCRWLL---PIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCL 196
+P + E L N+K P+QKY+I + KG DL+ACA TG+GKT FL PL +
Sbjct 158 SPPLDELLMENIKLASFTKPTPVQKYSIPIVTKGRDLMACAQTGSGKTGGFLFPLFT--- 214
Query 197 LHQQLFR------PHFAGSHA--QASPLAVVLCPTRELAIQINEQLARLVVGTTLSHSIW 248
+LFR P A S + P A+VL PTRELA QI E+ + + + +
Sbjct 215 ---ELFRSGPSPVPEKAQSFYSRKGYPSALVLAPTRELATQIFEEARKFTYRSWVRPCVV 271
Query 249 IGGVSATEQVREINQ 263
GG Q+RE+++
Sbjct 272 YGGAPIGNQMREVDR 286
> cel:Y71H2AM.19 hypothetical protein; K11594 ATP-dependent RNA
helicase [EC:3.6.4.13]
Length=643
Score = 76.6 bits (187), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 65/196 (33%), Positives = 92/196 (46%), Gaps = 29/196 (14%)
Query 62 PQRGDTKDEAAAAAAAAAAAAEADGDNEDELFDLSLTGGMFFDLLEQEAVCTCVGGDIEA 121
P RG +K E A D E ELF L+G + FD E E G D+
Sbjct 115 PSRGTSKWENRGAR---------DERIEQELFSGQLSG-INFDKYE-EIPVEATGDDV-- 161
Query 122 PEPAETLEQCSEGDSSLLAPRVVENLKNNLKCRWLLPIQKYAIAVAAKGLDLVACAATGT 181
P+P S L + EN+K R P+QKY+I G DL++CA TG+
Sbjct 162 PQPISLFSDLS------LHEWIEENIKTAGYDR-PTPVQKYSIPALQGGRDLMSCAQTGS 214
Query 182 GKTLAFLSPLVSKCL------LHQQLFRPHFAGSHAQASPLAVVLCPTRELAIQINEQLA 235
GKT AFL PLV+ L +H+ + +G + P A+VL PTREL++QI +
Sbjct 215 GKTAAFLVPLVNAILQDGPDAVHRSVTS---SGGRKKQYPSALVLSPTRELSLQIFNESR 271
Query 236 RLVVGTTLSHSIWIGG 251
+ T ++ ++ GG
Sbjct 272 KFAYRTPITSALLYGG 287
> xla:398019 ddx4, vasa, vlg1; DEAD (Asp-Glu-Ala-Asp) box polypeptide
4 (EC:3.6.4.13); K13982 probable ATP-dependent RNA helicase
DDX4 [EC:3.6.4.13]
Length=700
Score = 75.9 bits (185), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 63/224 (28%), Positives = 104/224 (46%), Gaps = 34/224 (15%)
Query 52 YGAYRGRS--------VSPQRGDTKDEAAAAAAAAAAAAEADGDNEDELFDLSLTGGMFF 103
YG Y+GR+ S + G+ KDE D ED +F G+ F
Sbjct 199 YGGYKGRNEEVGVESGKSQEEGNEKDEKPKKVTYIPPPPP---DGEDNIF-RQYQSGINF 254
Query 104 DLLEQEAVCTCVGGDIEAPEPAETLEQCSEGDSSLLAPRVVENLKNNL-KCRW--LLPIQ 160
D + E + G D+ P T E+ + + E L+ N+ + + L P+Q
Sbjct 255 DKYD-EILVDVTGKDV--PPAILTFEEAN----------LCETLRRNVARAGYVKLTPVQ 301
Query 161 KYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQ-ASPLAVV 219
K++I + G DL+ACA TG+GKT AFL P++S ++++ + A + Q P A++
Sbjct 302 KHSIPIIMAGRDLMACAQTGSGKTAAFLLPILS-YMMNEGI----TASQYLQLQEPEAII 356
Query 220 LCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREINQ 263
+ PTREL QI + GT + + GG+ +R++ +
Sbjct 357 IAPTRELINQIYLDARKFSYGTCVRPVVVYGGIQPVHAMRDVEK 400
> xla:397935 an3; ATP dependent RNA helicase (EC:3.6.4.13); K11594
ATP-dependent RNA helicase [EC:3.6.4.13]
Length=697
Score = 73.6 bits (179), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 41/113 (36%), Positives = 63/113 (55%), Gaps = 13/113 (11%)
Query 158 PIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQAS--- 214
P+QK+AI + + DL+ACA TG+GKT AFL P++S+ P A H Q +
Sbjct 246 PVQKHAIPIIIEKRDLMACAQTGSGKTAAFLLPILSQIYADG----PGDAMKHLQENGRY 301
Query 215 ------PLAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREI 261
PL++VL PTRELA+QI E+ + + + + GG +Q+R++
Sbjct 302 GRRKQFPLSLVLAPTRELAVQIYEEARKFAYRSRVRPCVVYGGADIGQQIRDL 354
> cel:B0414.6 glh-3; Germ-Line Helicase family member (glh-3)
Length=720
Score = 73.2 bits (178), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 51/169 (30%), Positives = 82/169 (48%), Gaps = 12/169 (7%)
Query 87 DNEDELFD-LSLTGGMFFDLLEQEAVCTCVGGDIEAPEPAETLEQCSEGDSSLLAPRVVE 145
D +E+F L + G FFD +V G +P T++ C S + +
Sbjct 258 DKMEEVFSMLKINAGDFFDKFFDASVQLVSRG-----QPV-TIQPCKSFSDSDIPQSMRR 311
Query 146 NLKNNLKCRWLLPIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPH 205
N++ R PIQ+Y + + A G D++ACA TG+GKT AFL P++S+ +L + L
Sbjct 312 NVERAGYTR-TTPIQQYTLPLVADGKDILACAQTGSGKTAAFLLPIMSRLILEKDL---- 366
Query 206 FAGSHAQASPLAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSA 254
G+ P ++L PTRELA QI + + + + GG++
Sbjct 367 NYGAEGGCYPRCIILTPTRELADQIYNEGRKFSYQSVMEIKPVYGGINV 415
> bbo:BBOV_IV001840 21.m02846; DEAD/DEAH box helicase (EC:3.6.1.-);
K11594 ATP-dependent RNA helicase [EC:3.6.4.13]
Length=609
Score = 72.4 bits (176), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 59/184 (32%), Positives = 88/184 (47%), Gaps = 18/184 (9%)
Query 87 DNEDELF---DLSLTGGMFFDLLEQEAVCTCVGGDIEAPEPAETLEQCSEGDSSLLAPRV 143
++E E+F D G+ F E V G ++ +P E E G LL
Sbjct 109 ESEKEVFGEMDTCTHAGINFGSYENIPV-EITGNQSQSIKPVEDFEN---GIHELL---- 160
Query 144 VENLKNNLKCRWL--LPIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCL-LHQQ 200
+ N L+ + PIQK++I V G DL+ACA TG+GKT AFL P+ + L
Sbjct 161 ---MVNILRVNYTKPTPIQKHSIPVIMAGRDLMACAQTGSGKTAAFLLPICTAMLKTGPP 217
Query 201 LFRPHFAGSHA-QASPLAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVR 259
RP + H+ QA P+ +VL PTRELA+Q + + + T + + GG Q+
Sbjct 218 ASRPMQSSYHSRQALPVCLVLSPTRELAMQTFTEARKFIYNTGIRAVVLYGGGEVRRQLY 277
Query 260 EINQ 263
E+ +
Sbjct 278 ELER 281
> ath:AT3G02065 DEAD/DEAH box helicase family protein
Length=505
Score = 72.0 bits (175), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 49/143 (34%), Positives = 72/143 (50%), Gaps = 12/143 (8%)
Query 121 APEPAETLEQCSEGDSSLLAPRVVENLKNNLKCRWLLPIQKYAIAVAAKGLDLVACAATG 180
P P T C L P+++ NL+ + PIQ AI A G L+A A TG
Sbjct 105 VPPPVLTFTSCG------LPPKLLLNLETA-GYDFPTPIQMQAIPAALTGKSLLASADTG 157
Query 181 TGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQASPLAVVLCPTRELAIQINEQLARLVVG 240
+GKT +FL P++S+C + H S + +PLA+VL PTREL +Q+ +Q L G
Sbjct 158 SGKTASFLVPIISRCTTYHS---EH--PSDQRRNPLAMVLAPTRELCVQVEDQAKMLGKG 212
Query 241 TTLSHSIWIGGVSATEQVREINQ 263
++ +GG + Q+ I Q
Sbjct 213 LPFKTALVVGGDPMSGQLYRIQQ 235
> hsa:54514 DDX4, MGC111074, VASA; DEAD (Asp-Glu-Ala-Asp) box
polypeptide 4 (EC:3.6.4.13); K13982 probable ATP-dependent RNA
helicase DDX4 [EC:3.6.4.13]
Length=690
Score = 71.6 bits (174), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 42/124 (33%), Positives = 66/124 (53%), Gaps = 7/124 (5%)
Query 143 VVENLKNNL-KCRW--LLPIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQ 199
+ + L NN+ K + L P+QKY+I + G DL+ACA TG+GKT AFL P+++ ++H
Sbjct 261 LCQTLNNNIAKAGYTKLTPVQKYSIPIILAGRDLMACAQTGSGKTAAFLLPILAH-MMHD 319
Query 200 QLFRPHFAGSHAQASPLAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVR 259
+ F P +++ PTREL QI + + GT + + GG +R
Sbjct 320 GITASRF---KELQEPECIIVAPTRELVNQIYLEARKFSFGTCVRAVVIYGGTQLGHSIR 376
Query 260 EINQ 263
+I Q
Sbjct 377 QIVQ 380
> mmu:26900 Ddx3y, 8030469F12Rik, D1Pas1-rs1, Dby; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 3, Y-linked (EC:3.6.4.13); K11594
ATP-dependent RNA helicase [EC:3.6.4.13]
Length=658
Score = 71.2 bits (173), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 36/111 (32%), Positives = 63/111 (56%), Gaps = 5/111 (4%)
Query 158 PIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQ-----QLFRPHFAGSHAQ 212
P+QK+AI + + DL+ACA TG+GKT AFL P++S+ + + + +
Sbjct 204 PVQKHAIPIIKEKRDLMACAQTGSGKTAAFLLPILSQIYTDGPGEALKAMKENGRYGRRK 263
Query 213 ASPLAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREINQ 263
P+++VL PTRELA+QI E+ + + + + GG +Q+R++ +
Sbjct 264 QYPISLVLAPTRELAVQIYEEARKFSYRSRVRPCVVYGGADTVQQIRDLER 314
> cel:Y54E10A.9 vbh-1; Vasa- and Belle-like Helicase family member
(vbh-1); K11594 ATP-dependent RNA helicase [EC:3.6.4.13]
Length=641
Score = 71.2 bits (173), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 53/165 (32%), Positives = 79/165 (47%), Gaps = 13/165 (7%)
Query 89 EDELFDLSLTGGMFFDLLEQEAVCTCVGGDIEAPEPAETLEQCSEGDSSLLAPRVVENLK 148
E LF + G+ FD + E + V GD P E + G P V+EN+
Sbjct 87 ESNLFHRT-DSGINFD--KYENIPVEVSGD-SVPAAIEHFNEAGFG------PAVMENVN 136
Query 149 NNLKCRWLLPIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLH--QQLFRPHF 206
+ + P+QK++I DL++CA TG+GKT AFL P++ L + P F
Sbjct 137 RSGYSK-PTPVQKHSIPTLLANRDLMSCAQTGSGKTAAFLLPIIQHILAGGPDMVKPPAF 195
Query 207 AGSHAQASPLAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGG 251
P A+VL PTRELAIQI+++ + + + +I GG
Sbjct 196 TNGRRTYYPCALVLSPTRELAIQIHKEATKFSYKSNIQTAILYGG 240
> xla:379975 ddx3x, MGC52935, pl10; DEAD (Asp-Glu-Ala-Asp) box
polypeptide 3, X-linked; K11594 ATP-dependent RNA helicase
[EC:3.6.4.13]
Length=697
Score = 70.9 bits (172), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 40/113 (35%), Positives = 62/113 (54%), Gaps = 13/113 (11%)
Query 158 PIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQAS--- 214
P+QK+AI + DL+ACA TG+GKT AFL P++S+ P A H + +
Sbjct 246 PVQKHAIPIIIGKRDLMACAQTGSGKTAAFLLPILSQIYADG----PGDAMKHLKDNGRY 301
Query 215 ------PLAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREI 261
PL++VL PTRELA+QI E+ + + + + GG +Q+R++
Sbjct 302 GRRKQFPLSLVLAPTRELAVQIYEEARKFAYRSRVRPCVVYGGADIGQQIRDL 354
> tgo:TGME49_026250 ATP-dependent RNA helicase, putative ; K11594
ATP-dependent RNA helicase [EC:3.6.4.13]
Length=734
Score = 70.5 bits (171), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 41/111 (36%), Positives = 58/111 (52%), Gaps = 6/111 (5%)
Query 158 PIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQ----- 212
PIQK +I G DL+ACA TG+GKT AFL P++++ L P A
Sbjct 255 PIQKNSIPTILSGRDLMACAQTGSGKTAAFLYPIIARMLQDGPPPLPQAAAGGGSGYRKP 314
Query 213 -ASPLAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREIN 262
A P+ +VL PTRELA+QI E+ + GT + GG Q+ +++
Sbjct 315 PAYPICLVLSPTRELAMQIYEEARKFQFGTGVRTVAVYGGSDVKRQLIDLD 365
> mmu:110957 D1Pas1, AU016353, Pl10; DNA segment, Chr 1, Pasteur
Institute 1 (EC:3.6.4.13); K11594 ATP-dependent RNA helicase
[EC:3.6.4.13]
Length=660
Score = 70.5 bits (171), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 36/111 (32%), Positives = 63/111 (56%), Gaps = 5/111 (4%)
Query 158 PIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQ-----QLFRPHFAGSHAQ 212
P+QK+AI + + DL+ACA TG+GKT AFL P++S+ + + + +
Sbjct 204 PVQKHAIPIIKEKRDLMACAQTGSGKTAAFLLPILSQIYTDGPGEALRAMKENGKYGRRK 263
Query 213 ASPLAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREINQ 263
P+++VL PTRELA+QI E+ + + + + GG +Q+R++ +
Sbjct 264 QYPISLVLAPTRELAVQIYEEARKFSYRSRVRPCVVYGGADIGQQIRDLER 314
> dre:566947 ddx3, fb74g09, wu:fb74g09, zgc:158804; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 3; K11594 ATP-dependent RNA helicase
[EC:3.6.4.13]
Length=709
Score = 69.7 bits (169), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 57/189 (30%), Positives = 88/189 (46%), Gaps = 33/189 (17%)
Query 89 EDELFDLSLTGGMF--FDLLEQEAVCTCVGGDIEAPEPAETLEQCSEGDSSLLAPRVVEN 146
E ELF S TG F +D + EA T G IE+ + + ++ N
Sbjct 190 EHELFSASNTGINFEKYDDIPVEATGTNSPGHIESFHDVD------------MGEIIMGN 237
Query 147 LKNNLKCRWLLPIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHF 206
+ + R P+QKYAI + DL+ACA TG+GKT AFL P++S+ P
Sbjct 238 ITLSRYTR-PTPVQKYAIPIIKTKRDLMACAQTGSGKTAAFLLPVLSQIYSEG----PGE 292
Query 207 AGSHAQAS--------------PLAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGV 252
A +AS P+++VL PTRELA+QI ++ + + + + GG
Sbjct 293 ALQATKASTQQENGKYVRRKQYPISLVLAPTRELALQIYDEARKFAYRSRVRPCVVYGGA 352
Query 253 SATEQVREI 261
+Q+R++
Sbjct 353 DIGQQIRDL 361
> mmu:13206 Ddx4, AV206478, Mvh, VASA; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 4 (EC:3.6.4.13); K13982 probable ATP-dependent
RNA helicase DDX4 [EC:3.6.4.13]
Length=728
Score = 69.7 bits (169), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 43/126 (34%), Positives = 65/126 (51%), Gaps = 11/126 (8%)
Query 143 VVENLKNNL-KCRW--LLPIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQ 199
+ + L NN+ K + L P+QKY+I + G DL+ACA TG+GKT AFL P+++
Sbjct 294 LCQTLNNNIAKAGYTKLTPVQKYSIPIVLAGRDLMACAQTGSGKTAAFLLPILA------ 347
Query 200 QLFRPHFAGSHAQA--SPLAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQ 257
+ R S + P +++ PTREL QI + + GT + + GG
Sbjct 348 HMMRDGITASRFKELQEPECIIVAPTRELINQIYLEARKFSFGTCVRAVVIYGGTQFGHS 407
Query 258 VREINQ 263
VR+I Q
Sbjct 408 VRQIVQ 413
> hsa:1654 DDX3X, DBX, DDX14, DDX3, HLP2; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 3, X-linked (EC:3.6.4.13); K11594 ATP-dependent
RNA helicase [EC:3.6.4.13]
Length=661
Score = 69.7 bits (169), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 36/111 (32%), Positives = 63/111 (56%), Gaps = 5/111 (4%)
Query 158 PIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQ-----QLFRPHFAGSHAQ 212
P+QK+AI + + DL+ACA TG+GKT AFL P++S+ + + + +
Sbjct 205 PVQKHAIPIIKEKRDLMACAQTGSGKTAAFLLPILSQIYSDGPGEALRAMKENGRYGRRK 264
Query 213 ASPLAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREINQ 263
P+++VL PTRELA+QI E+ + + + + GG +Q+R++ +
Sbjct 265 QYPISLVLAPTRELAVQIYEEARKFSYRSRVRPCVVYGGADIGQQIRDLER 315
> mmu:13205 Ddx3x, D1Pas1-rs2, Ddx3, Fin14; DEAD/H (Asp-Glu-Ala-Asp/His)
box polypeptide 3, X-linked (EC:3.6.4.13); K11594
ATP-dependent RNA helicase [EC:3.6.4.13]
Length=662
Score = 69.3 bits (168), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 36/111 (32%), Positives = 63/111 (56%), Gaps = 5/111 (4%)
Query 158 PIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQ-----QLFRPHFAGSHAQ 212
P+QK+AI + + DL+ACA TG+GKT AFL P++S+ + + + +
Sbjct 205 PVQKHAIPIIKEKRDLMACAQTGSGKTAAFLLPILSQIYADGPGEALRAMKENGRYGRRK 264
Query 213 ASPLAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREINQ 263
P+++VL PTRELA+QI E+ + + + + GG +Q+R++ +
Sbjct 265 QYPISLVLAPTRELAVQIYEEARKFSYRSRVRPCVVYGGAEIGQQIRDLER 315
> hsa:8653 DDX3Y, DBY; DEAD (Asp-Glu-Ala-Asp) box polypeptide
3, Y-linked (EC:3.6.4.13); K11594 ATP-dependent RNA helicase
[EC:3.6.4.13]
Length=660
Score = 68.9 bits (167), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 36/111 (32%), Positives = 62/111 (55%), Gaps = 5/111 (4%)
Query 158 PIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQ-----QLFRPHFAGSHAQ 212
P+QK+AI + DL+ACA TG+GKT AFL P++S+ + + + +
Sbjct 203 PVQKHAIPIIKGKRDLMACAQTGSGKTAAFLLPILSQIYTDGPGEALKAVKENGRYGRRK 262
Query 213 ASPLAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREINQ 263
P+++VL PTRELA+QI E+ + + + + GG +Q+R++ +
Sbjct 263 QYPISLVLAPTRELAVQIYEEARKFSYRSRVRPCVVYGGADIGQQIRDLER 313
> dre:321948 ddx46, fb39a03, wu:fb39a03; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 46 (EC:3.6.4.13); K12811 ATP-dependent RNA
helicase DDX46/PRP5 [EC:3.6.4.13]
Length=1018
Score = 68.6 bits (166), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 48/142 (33%), Positives = 71/142 (50%), Gaps = 15/142 (10%)
Query 122 PEPAETLEQCSEGDSSLLAPRVVENLKNNLKCRWLLPIQKYAIAVAAKGLDLVACAATGT 181
P+P +T QC ++ +V+ LK + PIQ AI G DL+ A TG+
Sbjct 336 PKPIKTWVQCG------ISMKVLNALKKH-NYEKPTPIQAQAIPAIMSGRDLIGIAKTGS 388
Query 182 GKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQASPLAVVLCPTRELAIQINEQLARLVVGT 241
GKT+AFL P+ + +L Q RP PLAV++ PTRELA+QI ++ +
Sbjct 389 GKTIAFLLPMF-RHILDQ---RP----VGEAEGPLAVIMTPTRELALQITKECKKFSKSL 440
Query 242 TLSHSIWIGGVSATEQVREINQ 263
L GG +EQ+ E+ +
Sbjct 441 ALRVVCVYGGTGISEQIAELKR 462
> bbo:BBOV_II004470 18.m06373; p68-like protein; K12823 ATP-dependent
RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=529
Score = 68.2 bits (165), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 51/160 (31%), Positives = 73/160 (45%), Gaps = 16/160 (10%)
Query 104 DLLEQEAVCTCVGGDIEAPEPAETLEQCSEGDSSLLAPRVVENLKNNLKCRWLLPIQKYA 163
D + +E T + G + P+P + E S D L A R PIQ
Sbjct 87 DRVRKEREITIIAGR-DVPKPVVSFEHTSFPDYILKAIRAAGFTAPT-------PIQVQG 138
Query 164 IAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQASPLAVVLCPT 223
+A G D++ A TG+GKTLAFL P V + Q L RP P+ +VL PT
Sbjct 139 WPIALSGRDVIGIAETGSGKTLAFLLPAVVH-INAQHLLRP-------GDGPIVLVLAPT 190
Query 224 RELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREINQ 263
REL QI +Q + + + S+ GGV +Q+ E+ +
Sbjct 191 RELVEQIRQQCVQFGASSRIKSSVAYGGVPKRQQMYELKR 230
> mmu:67848 Ddx55, 2810021H22Rik, MGC143677, MGC143678, mKIAA1595;
DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 (EC:3.6.4.13);
K14809 ATP-dependent RNA helicase DDX55/SPB4 [EC:3.6.4.13]
Length=596
Score = 68.2 bits (165), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 50/141 (35%), Positives = 72/141 (51%), Gaps = 14/141 (9%)
Query 128 LEQCSEGD-SSL---LAPRVVENLKNNLKCRWLLPIQKYAIAVAAKGLDLVACAATGTGK 183
+E +EG SL L PRV+ L+ L + P+Q I + K D+ A A TG+GK
Sbjct 1 MEHVTEGAWESLQVPLHPRVLGALRE-LGFPHMTPVQSATIPLFMKNKDVAAEAVTGSGK 59
Query 184 TLAFLSPLVSKCLLHQQLFRPHFAGSHAQASPLAVVLCPTRELAIQINEQLARLVVGT-T 242
TLAF+ P++ L ++ + + G A+V+ PTRELAIQI+E L+
Sbjct 60 TLAFVIPILEILLRREEKLKKNQVG--------AIVITPTRELAIQIDEVLSHFTKHFPQ 111
Query 243 LSHSIWIGGVSATEQVREINQ 263
S +WIGG + E V Q
Sbjct 112 FSQILWIGGRNPGEDVERFKQ 132
> sce:YLL008W DRS1; Drs1p (EC:3.6.1.-); K13181 ATP-dependent RNA
helicase DDX27 [EC:3.6.4.13]
Length=752
Score = 68.2 bits (165), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 40/103 (38%), Positives = 59/103 (57%), Gaps = 15/103 (14%)
Query 158 PIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQASPLA 217
PIQ I +A G D++A A TG+GKT AF+ P++ + L++P AS
Sbjct 256 PIQSATIPIALLGKDIIAGAVTGSGKTAAFMIPIIERL-----LYKP-----AKIASTRV 305
Query 218 VVLCPTRELAIQ---INEQLARLVVGTTLSHSIWIGGVSATEQ 257
+VL PTRELAIQ + +Q+AR V G T + +GG++ +Q
Sbjct 306 IVLLPTRELAIQVADVGKQIARFVSGITF--GLAVGGLNLRQQ 346
> dre:30116 pl10, etID309900.24, p110, p110a, pl10a, wu:fb43h11,
wu:fy72b06; pl10; K11594 ATP-dependent RNA helicase [EC:3.6.4.13]
Length=688
Score = 67.8 bits (164), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 54/184 (29%), Positives = 87/184 (47%), Gaps = 24/184 (13%)
Query 89 EDELFDLSLTGGMF--FDLLEQEAVCTCVGGDIEAPEPAETLEQCSEGDSSLLAPRVVEN 146
E ELF S TG F +D + EA P+P + G+ ++ N
Sbjct 175 EHELFSGSNTGINFEKYDDIPVEATGH------NGPQPIDRFHDLEMGEI------IMGN 222
Query 147 LKNNLKCRWLLPIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHF 206
+ + R P+QK+AI + DL+ACA TG+GKT AFL P++S+
Sbjct 223 INLSRYTR-PTPVQKHAIPIIKSKRDLMACAQTGSGKTAAFLLPVLSQIYTDGPGEALQA 281
Query 207 AGSHAQAS---------PLAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQ 257
A + AQ + P+++VL PTRELA+QI ++ + + + + GG +Q
Sbjct 282 AKNSAQENGKYGRRKQYPISLVLAPTRELALQIYDEARKFSYRSHVRPCVVYGGADIGQQ 341
Query 258 VREI 261
+R++
Sbjct 342 IRDL 345
> dre:30263 vasa, MGC158535, fi24g05, vas, vlg, wu:fi24g05, zgc:109812,
zgc:158535; vasa homolog (EC:3.6.4.13); K13982 probable
ATP-dependent RNA helicase DDX4 [EC:3.6.4.13]
Length=716
Score = 67.8 bits (164), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 37/106 (34%), Positives = 57/106 (53%), Gaps = 8/106 (7%)
Query 158 PIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQA--SP 215
P+QK+ I + + G DL+ACA TG+GKT AFL P++ Q+ A S P
Sbjct 302 PVQKHGIPIISAGRDLMACAQTGSGKTAAFLLPIL------QRFMTDGVAASKFSEIQEP 355
Query 216 LAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREI 261
A+++ PTREL QI + + GT + + GG++ +RE+
Sbjct 356 EAIIVAPTRELINQIYLEARKFAYGTCVRPVVVYGGINTGYTIREV 401
> hsa:57696 DDX55, FLJ16577, KIAA1595, MGC33209; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 55 (EC:3.6.4.13); K14809 ATP-dependent
RNA helicase DDX55/SPB4 [EC:3.6.4.13]
Length=600
Score = 67.0 bits (162), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 47/142 (33%), Positives = 73/142 (51%), Gaps = 14/142 (9%)
Query 128 LEQCSEGD-SSL---LAPRVVENLKNNLKCRWLLPIQKYAIAVAAKGLDLVACAATGTGK 183
+E +EG SL L P+V+ L+ L ++ P+Q I + + D+ A A TG+GK
Sbjct 1 MEHVTEGSWESLPVPLHPQVLGALRE-LGFPYMTPVQSATIPLFMRNKDVAAEAVTGSGK 59
Query 184 TLAFLSPLVSKCLLHQQLFRPHFAGSHAQASPLAVVLCPTRELAIQINEQLARLVVGT-T 242
TLAF+ P++ L ++ + G A+++ PTRELAIQI+E L+
Sbjct 60 TLAFVIPILEILLRREEKLKKSQVG--------AIIITPTRELAIQIDEVLSHFTKHFPE 111
Query 243 LSHSIWIGGVSATEQVREINQK 264
S +WIGG + E V Q+
Sbjct 112 FSQILWIGGRNPGEDVERFKQQ 133
> cel:H20J04.4 hypothetical protein; K14778 ATP-dependent RNA
helicase DDX49/DBP8 [EC:3.6.4.13]
Length=561
Score = 66.6 bits (161), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 42/107 (39%), Positives = 59/107 (55%), Gaps = 13/107 (12%)
Query 158 PIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQASPLA 217
P+Q I +G D++ CA TGTGKTLAF P++ K + P+ G +A
Sbjct 114 PVQAACIPKILEGSDILGCARTGTGKTLAFAIPILQKLSVD-----PY--GIYA------ 160
Query 218 VVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREINQK 264
++L PTRELA QI EQ L TL S+ +GG S Q RE++++
Sbjct 161 LILTPTRELAFQIAEQFTALGKPITLKCSVIVGGRSLIHQARELSER 207
> ath:AT3G22310 PMH1; PMH1 (PUTATIVE MITOCHONDRIAL RNA HELICASE
1); ATP-dependent helicase/ DNA binding / RNA binding
Length=610
Score = 66.2 bits (160), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 43/125 (34%), Positives = 64/125 (51%), Gaps = 12/125 (9%)
Query 139 LAPRVVENLKNNLKCRWLLPIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLH 198
++P +V+ LK L PIQK + A +G D++ A TGTGKTLAF P++ K +
Sbjct 123 ISPEIVKALKGR-GIEKLFPIQKAVLEPAMEGRDMIGRARTGTGKTLAFGIPIIDKII-- 179
Query 199 QQLFRPHFAGSHAQA-SPLAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQ 257
F H + +P +VL PTRELA Q+ ++ +L GG +Q
Sbjct 180 ------KFNAKHGRGKNPQCLVLAPTRELARQVEKEFRE--SAPSLDTICLYGGTPIGQQ 231
Query 258 VREIN 262
+RE+N
Sbjct 232 MRELN 236
> cel:T21G5.3 glh-1; Germ-Line Helicase family member (glh-1)
Length=763
Score = 65.9 bits (159), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 34/96 (35%), Positives = 54/96 (56%), Gaps = 4/96 (4%)
Query 158 PIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQASPLA 217
PIQ+YA+ + +G D++ACA TG+GKT AFL P++++ + L G + P
Sbjct 366 PIQQYALPLVHQGYDIMACAQTGSGKTAAFLLPIMTRLIDDNNLNTAGEGGCY----PRC 421
Query 218 VVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVS 253
++L PTRELA QI + + T + GG++
Sbjct 422 IILTPTRELADQIYNEGRKFAYQTMMEIKPVYGGLA 457
> dre:378844 ddx27, cb843; DEAD (Asp-Glu-Ala-Asp) box polypeptide
27 (EC:3.6.4.13); K13181 ATP-dependent RNA helicase DDX27
[EC:3.6.4.13]
Length=776
Score = 65.5 bits (158), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 38/100 (38%), Positives = 57/100 (57%), Gaps = 10/100 (10%)
Query 158 PIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQASPLA 217
PIQK + V G D+ ACAATGTGKT AF+ P++ + +++P Q + +
Sbjct 226 PIQKACVPVGLLGKDICACAATGTGKTAAFMLPVLERL-----IYKPR----ETQVTRV- 275
Query 218 VVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQ 257
+VL PTREL IQ++ +L TT+S + +GG+ Q
Sbjct 276 LVLVPTRELGIQVHTVARQLAQFTTISTCLAVGGLDLKSQ 315
> cel:Y71G12B.8 hypothetical protein; K13181 ATP-dependent RNA
helicase DDX27 [EC:3.6.4.13]
Length=763
Score = 65.5 bits (158), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 40/100 (40%), Positives = 54/100 (54%), Gaps = 10/100 (10%)
Query 158 PIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQASPLA 217
PIQ+ I VA G D+ ACAATGTGKT AF+ P +L + ++RP A +
Sbjct 173 PIQQACIPVALTGKDICACAATGTGKTAAFVLP-----ILERMIYRPKGA-----SCTRV 222
Query 218 VVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQ 257
+VL PTRELAIQ+ + +L L + GG+ Q
Sbjct 223 LVLVPTRELAIQVFQVFRKLSTFIQLEVCLCAGGLDLKAQ 262
> ath:AT1G55150 DEAD box RNA helicase, putative (RH20); K12823
ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=501
Score = 65.1 bits (157), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 37/106 (34%), Positives = 58/106 (54%), Gaps = 8/106 (7%)
Query 158 PIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQASPLA 217
PIQ +A KG DL+ A TG+GKTL++L P + ++ Q H G P+
Sbjct 124 PIQSQGWPMAMKGRDLIGIAETGSGKTLSYLLPAI--VHVNAQPMLAHGDG------PIV 175
Query 218 VVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREINQ 263
+VL PTRELA+QI ++ ++ + + + GGV QVR++ +
Sbjct 176 LVLAPTRELAVQIQQEASKFGSSSKIKTTCIYGGVPKGPQVRDLQK 221
> sce:YJL033W HCA4, DBP4, ECM24; Hca4p (EC:3.6.1.-); K14776 ATP-dependent
RNA helicase DDX10/DBP4 [EC:3.6.4.13]
Length=770
Score = 65.1 bits (157), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 40/111 (36%), Positives = 62/111 (55%), Gaps = 9/111 (8%)
Query 156 LLPIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQASP 215
L IQ +I V+ +G D++A A TG+GKTLAFL P++ K L+R +
Sbjct 64 LTEIQADSIPVSLQGHDVLAAAKTGSGKTLAFLVPVIEK------LYREKWTEFDGLG-- 115
Query 216 LAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREINQKQI 266
A+++ PTRELA+QI E L ++ T+ S + IGG ++ I++ I
Sbjct 116 -ALIISPTRELAMQIYEVLTKIGSHTSFSAGLVIGGKDVKFELERISRINI 165
> ath:AT3G01540 DRH1; DRH1 (DEAD BOX RNA HELICASE 1); ATP-dependent
RNA helicase/ ATPase
Length=619
Score = 65.1 bits (157), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 38/106 (35%), Positives = 57/106 (53%), Gaps = 9/106 (8%)
Query 158 PIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQASPLA 217
PIQ + +A +G D+VA A TG+GKTL +L P LH Q R + ++ P
Sbjct 183 PIQAQSWPIAMQGRDIVAIAKTGSGKTLGYLIP----GFLHLQRIR-----NDSRMGPTI 233
Query 218 VVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREINQ 263
+VL PTRELA QI E+ + + +S + GG Q+R++ +
Sbjct 234 LVLSPTRELATQIQEEAVKFGRSSRISCTCLYGGAPKGPQLRDLER 279
> xla:380261 ddx42, MGC54025; DEAD (Asp-Glu-Ala-Asp) box polypeptide
42 (EC:3.6.4.13); K12835 ATP-dependent RNA helicase DDX42
[EC:3.6.4.13]
Length=947
Score = 65.1 bits (157), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 36/106 (33%), Positives = 53/106 (50%), Gaps = 8/106 (7%)
Query 158 PIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQASPLA 217
PIQ I VA G D++ A TG+GKT AF+ P++ + ++L P+A
Sbjct 275 PIQCQGIPVALSGRDMIGIAKTGSGKTAAFIWPILVHIMDQKEL--------QPADGPIA 326
Query 218 VVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREINQ 263
V++CPTREL QI+ + R L GG S EQ + + +
Sbjct 327 VIVCPTRELCQQIHSECKRFGKAYNLRSVAVYGGGSMWEQAKALQE 372
> cpv:cgd8_800 Prp5p C terminal KH. eIF4A-1-family RNA SFII helicase
Length=934
Score = 64.7 bits (156), Expect = 3e-10, Method: Composition-based stats.
Identities = 37/110 (33%), Positives = 60/110 (54%), Gaps = 5/110 (4%)
Query 157 LPIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQAS-- 214
PIQ +I + G D++ A TG+GKTLA++ PL+ L+ P A Q +
Sbjct 246 FPIQMQSIPILMSGYDMIGNAETGSGKTLAYILPLIRHVLVQSNNNYPFNAEMDIQINKN 305
Query 215 ---PLAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREI 261
A+++ PTRELA+Q+ +Q +L L+ +I GG+S + Q+ +I
Sbjct 306 TNLARAMIIIPTRELALQVYKQTTQLANLVDLTTNIICGGLSISHQLNKI 355
> xla:398189 RNA helicase II/Gu
Length=759
Score = 64.3 bits (155), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 50/132 (37%), Positives = 65/132 (49%), Gaps = 14/132 (10%)
Query 133 EGDSSLLAPRVVENLKNNLKCR---WLLPIQKYAIAVAAKGLDLVACAATGTGKTLAFLS 189
+GD S P E +KN L+ + +L PIQ A G D+V A TGTGKT +F
Sbjct 178 DGDFSKF-PLSKETIKN-LQAKGVSYLFPIQSKTFHTAYSGKDVVVQARTGTGKTFSFAI 235
Query 190 PLVSKCLLHQQLFRPHFAGSHAQASPLAVVLCPTRELAIQINEQLARLVVGTTLSHSIWI 249
PLV K QQ P G +P ++L PTRELAIQI ++ + L S +
Sbjct 236 PLVEKLNEDQQ---PLARGR----APRVIILTPTRELAIQITNEIRSIT--KKLKVSCFY 286
Query 250 GGVSATEQVREI 261
GG +QV I
Sbjct 287 GGTPYQQQVFAI 298
> xla:444634 MGC84147 protein
Length=450
Score = 64.3 bits (155), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 36/106 (33%), Positives = 53/106 (50%), Gaps = 8/106 (7%)
Query 158 PIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQASPLA 217
PIQ I VA G D++ A TG+GKT AF+ P++ + ++L P+A
Sbjct 276 PIQCQGIPVALSGRDMIGIAKTGSGKTAAFIWPILVHIMDQKEL--------QPGDGPIA 327
Query 218 VVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREINQ 263
V++CPTREL QI+ + R L GG S EQ + + +
Sbjct 328 VIVCPTRELCQQIHNECKRFGKAYNLRSVAVYGGGSMWEQAKALQE 373
> pfa:PFL2475w DEAD/DEAH box helicase, putative; K13181 ATP-dependent
RNA helicase DDX27 [EC:3.6.4.13]
Length=717
Score = 63.9 bits (154), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 34/108 (31%), Positives = 66/108 (61%), Gaps = 4/108 (3%)
Query 159 IQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCL--LHQQLFRPHFAGSHAQASPL 216
IQ+ I +A +G ++A + TG+GKTLAF+ P++ + L ++ ++ R + GS+
Sbjct 113 IQRDVIPLALEGKSILANSETGSGKTLAFVLPILERLLQSVNIKMRRNNMKGSYNITK-- 170
Query 217 AVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREINQK 264
A++L PTREL++Q + + L T+++S++ GG+ +Q E ++
Sbjct 171 ALILLPTRELSLQCYDVIRSLTKYVTITYSLFCGGIDIKQQEYEFKKR 218
> hsa:83479 DDX59, DKFZp564B1023, ZNHIT5; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 59 (EC:3.6.4.13)
Length=619
Score = 63.9 bits (154), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 60/207 (28%), Positives = 91/207 (43%), Gaps = 39/207 (18%)
Query 61 SPQRGDTKDEAAAAAAAAAAAAEADGDNEDELFDLSLTGGMFFDLLEQEAVCTCVGGDIE 120
+PQ+ D++ E+ A+ ++ F L+L +L +Q + V G E
Sbjct 150 NPQKADSEPESPLNASYVY---------KEHPFILNLQEDQIENLKQQLGIL--VQGQ-E 197
Query 121 APEPAETLEQCSEGDSSLLAPRVVENLKNNLK---CRWLLPIQKYAIAVAAKGLDLVACA 177
P E CS + E L +NLK PIQ I V G D++A A
Sbjct 198 VTRPIIDFEHCS----------LPEVLNHNLKKSGYEVPTPIQMQMIPVGLLGRDILASA 247
Query 178 ATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQASPLAVVLCPTRELAIQINEQLARL 237
TG+GKT AFL P++ + L + +P A++L PTRELAIQI Q L
Sbjct 248 DTGSGKTAAFLLPVIMRALFESK-------------TPSALILTPTRELAIQIERQAKEL 294
Query 238 VVGTT-LSHSIWIGGVSATEQVREINQ 263
+ G + + +GG+ Q+ + Q
Sbjct 295 MSGLPRMKTVLLVGGLPLPPQLYRLQQ 321
> ath:AT3G22330 PMH2; PMH2 (putative mitochondrial RNA helicase
2); ATP binding / ATP-dependent helicase/ helicase/ nucleic
acid binding
Length=616
Score = 63.5 bits (153), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 40/125 (32%), Positives = 65/125 (52%), Gaps = 12/125 (9%)
Query 139 LAPRVVENLKNNLKCRWLLPIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLH 198
++P +V+ L + L PIQK + A +G D++ A TGTGKTLAF P++ K +
Sbjct 111 ISPEIVKALSSK-GIEKLFPIQKAVLEPAMEGRDMIGRARTGTGKTLAFGIPIIDKII-- 167
Query 199 QQLFRPHFAGSHAQA-SPLAVVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQ 257
+ H + +PL +VL PTRELA Q+ ++ +L GG +Q
Sbjct 168 ------KYNAKHGRGRNPLCLVLAPTRELARQVEKEFRE--SAPSLDTICLYGGTPIGQQ 219
Query 258 VREIN 262
+R+++
Sbjct 220 MRQLD 224
> cel:F53H1.1 hypothetical protein; K12811 ATP-dependent RNA helicase
DDX46/PRP5 [EC:3.6.4.13]
Length=970
Score = 63.5 bits (153), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 46/147 (31%), Positives = 71/147 (48%), Gaps = 19/147 (12%)
Query 119 IEAPEPAETLEQCSEGDSSLLAPRVVENLKNNLKCRWLLP--IQKYAIAVAAKGLDLVAC 176
I+ P+P +T QC + +++ LK K + P IQ AI G D++
Sbjct 297 IDCPKPIKTWAQCG------VNLKMMNVLK---KFEYSKPTSIQAQAIPSIMSGRDVIGI 347
Query 177 AATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQASPLAVVLCPTRELAIQINEQLAR 236
A TG+GKTLAFL P+ L +L P+AV+L PTRELA+Q ++ +
Sbjct 348 AKTGSGKTLAFLLPMFRHILDQPEL--------EEGDGPIAVILAPTRELAMQTYKEANK 399
Query 237 LVVGTTLSHSIWIGGVSATEQVREINQ 263
L + GGV +EQ+ ++ +
Sbjct 400 FAKPLGLKVACTYGGVGISEQIADLKR 426
> mmu:72047 Ddx42, 1810047H21Rik, AW319508, AW556242, B430002H05Rik,
RHELP, RNAHP, SF3b125; DEAD (Asp-Glu-Ala-Asp) box polypeptide
42 (EC:3.6.4.13); K12835 ATP-dependent RNA helicase
DDX42 [EC:3.6.4.13]
Length=929
Score = 63.5 bits (153), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 35/106 (33%), Positives = 53/106 (50%), Gaps = 8/106 (7%)
Query 158 PIQKYAIAVAAKGLDLVACAATGTGKTLAFLSPLVSKCLLHQQLFRPHFAGSHAQASPLA 217
PIQ + VA G D++ A TG+GKT AF+ P++ + ++L P+A
Sbjct 278 PIQCQGVPVALSGRDMIGIAKTGSGKTAAFIWPMLIHIMDQKEL--------EPGDGPIA 329
Query 218 VVLCPTRELAIQINEQLARLVVGTTLSHSIWIGGVSATEQVREINQ 263
V++CPTREL QI+ + R L GG S EQ + + +
Sbjct 330 VIVCPTRELCQQIHAECKRFGKAYNLRSVAVYGGGSMWEQAKALQE 375
Lambda K H
0.314 0.128 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9890911460
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40