bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_3342_orf2
Length=97
Score E
Sequences producing significant alignments: (Bits) Value
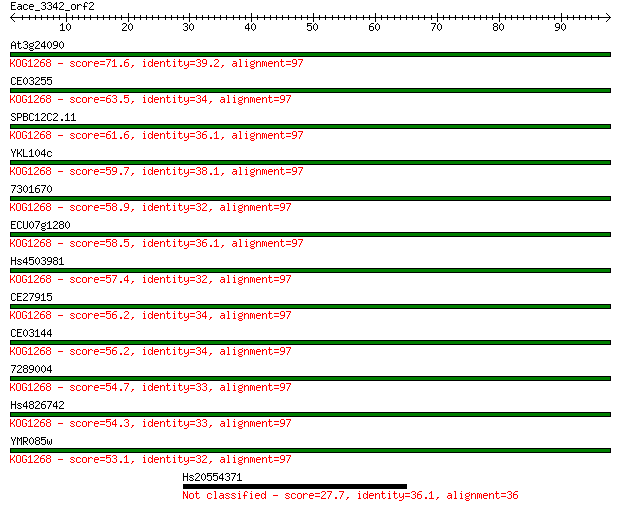
At3g24090 71.6 4e-13
CE03255 63.5 8e-11
SPBC12C2.11 61.6 4e-10
YKL104c 59.7 1e-09
7301670 58.9 2e-09
ECU07g1280 58.5 3e-09
Hs4503981 57.4 6e-09
CE27915 56.2 1e-08
CE03144 56.2 1e-08
7289004 54.7 5e-08
Hs4826742 54.3 5e-08
YMR085w 53.1 1e-07
Hs20554371 27.7 5.6
> At3g24090
Length=691
Score = 71.6 bits (174), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 38/97 (39%), Positives = 56/97 (57%), Gaps = 2/97 (2%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYA 60
CGV++NAG E+ VASTKA+TSQ+ V+ +++ + S + R A++D L LP
Sbjct 468 CGVHINAGAEIGVASTKAYTSQIVVMVMLALAIGSDTISSQ--KRREAIIDGLLDLPYKV 525
Query 61 GMTLNAHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
L K +A+ L D Q+L V G+G+ Y ALE
Sbjct 526 KEVLKLDDEMKDLAQLLIDEQSLLVFGRGYNYATALE 562
> CE03255
Length=710
Score = 63.5 bits (153), Expect = 8e-11, Method: Composition-based stats.
Identities = 33/97 (34%), Positives = 58/97 (59%), Gaps = 2/97 (2%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYA 60
CG+++NAG E+ VASTKA+TSQ+ L L + ++ S+ R + ++DAL+ LP+
Sbjct 488 CGIHINAGPEIGVASTKAYTSQIVSLLLFALTISEDRISK--MKRRAEIIDALNNLPILI 545
Query 61 GMTLNAHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
L+ +IAE++ ++L ++G+G + LE
Sbjct 546 RDVLDLDDQVLKIAEQIYKDKSLLIMGRGLNFATCLE 582
> SPBC12C2.11
Length=696
Score = 61.6 bits (148), Expect = 4e-10, Method: Composition-based stats.
Identities = 35/98 (35%), Positives = 59/98 (60%), Gaps = 3/98 (3%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYA 60
CGV++NAG E+ VASTKA+TSQ L L++ + ++ S +R + ++D L +
Sbjct 476 CGVHINAGPEICVASTKAYTSQYVALVLMALYLSRD--SVSRLERRNEIIDGLAEIGEKV 533
Query 61 GMTLNAHGACKRIA-EKLKDAQTLFVLGKGFGYPVALE 97
TL+ + A K+ A E+L + + ++G+G+ Y ALE
Sbjct 534 QETLHLNAAIKQTAIEQLINKDKMLIIGRGYHYATALE 571
> YKL104c
Length=717
Score = 59.7 bits (143), Expect = 1e-09, Method: Composition-based stats.
Identities = 37/100 (37%), Positives = 56/100 (56%), Gaps = 7/100 (7%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYS--DRCSALLDALHRLPV 58
CGV++NAG E+ VASTKA+TSQ L + F +L + S DR ++ L +P
Sbjct 491 CGVHINAGPEIGVASTKAYTSQYIALVM----FALSLSDDRVSKIDRRIEIIQGLKLIPG 546
Query 59 YAGMTLNAHGACKRI-AEKLKDAQTLFVLGKGFGYPVALE 97
L K++ A +LKD ++L +LG+G+ + ALE
Sbjct 547 QIKQVLKLEPRIKKLCATELKDQKSLLLLGRGYQFAAALE 586
> 7301670
Length=683
Score = 58.9 bits (141), Expect = 2e-09, Method: Composition-based stats.
Identities = 31/97 (31%), Positives = 57/97 (58%), Gaps = 2/97 (2%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYA 60
CGV++NAG E+ VASTKA+TSQ L + + ++ S + R ++D L +L +
Sbjct 462 CGVHINAGPEIGVASTKAYTSQFISLVMFALVMSEDRLSLQ--QRRLEIIDGLSQLDEHI 519
Query 61 GMTLNAHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
L + +++A++L + ++L ++G+GF + LE
Sbjct 520 RTVLKLNSQVQQLAKELYEHKSLLIMGRGFNFATCLE 556
> ECU07g1280
Length=699
Score = 58.5 bits (140), Expect = 3e-09, Method: Composition-based stats.
Identities = 35/99 (35%), Positives = 54/99 (54%), Gaps = 5/99 (5%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFW-QNLRSEEYSDRCSALLDALHRLPVY 59
CGV++NAG E+ VASTKA+TSQ L LV+ QNL + R +++ L +
Sbjct 478 CGVHINAGPEIGVASTKAYTSQYIALVLVALQLSDQNLVKQA---RRREIMEGLKNISSQ 534
Query 60 AGMTLNAHGACKRIAE-KLKDAQTLFVLGKGFGYPVALE 97
L + K +A +KD +L ++G+G+ YP +E
Sbjct 535 INRVLELSTSVKSLANGPMKDDASLLLIGRGYQYPTCME 573
> Hs4503981
Length=681
Score = 57.4 bits (137), Expect = 6e-09, Method: Composition-based stats.
Identities = 31/97 (31%), Positives = 54/97 (55%), Gaps = 2/97 (2%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYA 60
CGV++NAG E+ VASTKA+TSQ L + + + S + +R ++ L RLP
Sbjct 460 CGVHINAGPEIGVASTKAYTSQFVSLVMFALMMCDDRISMQ--ERRKEIMLGLKRLPDLI 517
Query 61 GMTLNAHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
L+ +++A +L +++ ++G+G+ Y LE
Sbjct 518 KEVLSMDDEIQKLATELYHQKSVLIMGRGYHYATCLE 554
> CE27915
Length=712
Score = 56.2 bits (134), Expect = 1e-08, Method: Composition-based stats.
Identities = 33/99 (33%), Positives = 53/99 (53%), Gaps = 6/99 (6%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYS--DRCSALLDALHRLPV 58
CG+++NAG E+ VASTKA+TSQ+ L + F L + S R ++DAL+ LP
Sbjct 490 CGIHINAGPEIGVASTKAYTSQILSLLM----FALTLSDDRISMAKRREEIIDALNDLPE 545
Query 59 YAGMTLNAHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
L IA+++ ++L ++G+G + LE
Sbjct 546 LIREVLQLDEKVLDIAKQIYKEKSLLIMGRGLNFATCLE 584
> CE03144
Length=725
Score = 56.2 bits (134), Expect = 1e-08, Method: Composition-based stats.
Identities = 33/99 (33%), Positives = 53/99 (53%), Gaps = 6/99 (6%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYS--DRCSALLDALHRLPV 58
CG+++NAG E+ VASTKA+TSQ+ L + F L + S R ++DAL+ LP
Sbjct 503 CGIHINAGPEIGVASTKAYTSQILSLLM----FALTLSDDRISMAKRREEIIDALNDLPE 558
Query 59 YAGMTLNAHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
L IA+++ ++L ++G+G + LE
Sbjct 559 LIREVLQLDEKVLDIAKQIYKEKSLLIMGRGLNFATCLE 597
> 7289004
Length=694
Score = 54.7 bits (130), Expect = 5e-08, Method: Composition-based stats.
Identities = 32/97 (32%), Positives = 52/97 (53%), Gaps = 2/97 (2%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYA 60
CGV++NAG E+ VASTKA+TSQ L + + ++ S + R +L AL +L
Sbjct 473 CGVHINAGPEIGVASTKAYTSQFISLVMFALVMSEDRLSLQ--QRRLEILQALSKLADQI 530
Query 61 GMTLNAHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
L K +A+ L ++L ++G+G+ + LE
Sbjct 531 RDVLQLDSKVKELAKDLYQHKSLLIMGRGYNFATCLE 567
> Hs4826742
Length=682
Score = 54.3 bits (129), Expect = 5e-08, Method: Composition-based stats.
Identities = 32/97 (32%), Positives = 51/97 (52%), Gaps = 2/97 (2%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYA 60
CGV++NAG E+ VASTKA+TSQ L + ++ S + +R ++ L LP
Sbjct 461 CGVHINAGPEIGVASTKAYTSQFISLVMFGLMMSEDRISLQ--NRRQEIIRGLRSLPELI 518
Query 61 GMTLNAHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
L+ +A +L ++L V+G+G+ Y LE
Sbjct 519 KEVLSLEEKIHDLALELYTQRSLLVMGRGYNYATCLE 555
> YMR085w
Length=432
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 31/98 (31%), Positives = 52/98 (53%), Gaps = 3/98 (3%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYA 60
CGV+ N G E +A+TK++TSQ L +++ W ++L S+ +R ++ AL +P
Sbjct 206 CGVHTNTGPEKGIATTKSYTSQYIALVMIALWMSEDLVSK--IERRKEIIQALTIVPSQI 263
Query 61 GMTLNAHGACKRIAE-KLKDAQTLFVLGKGFGYPVALE 97
L + + KLK T +LG+G+ + ALE
Sbjct 264 KEVLELEPLIIELCDKKLKQHDTFLLLGRGYQFASALE 301
> Hs20554371
Length=576
Score = 27.7 bits (60), Expect = 5.6, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 19/36 (52%), Gaps = 0/36 (0%)
Query 29 VSSWFWQNLRSEEYSDRCSALLDALHRLPVYAGMTL 64
++SWF R E + RCS + HR P + G+ L
Sbjct 112 LASWFCSGRRLEMTAYRCSHMKFLTHREPSHFGLDL 147
Lambda K H
0.322 0.133 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1198045380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40