bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_3287_orf3
Length=66
Score E
Sequences producing significant alignments: (Bits) Value
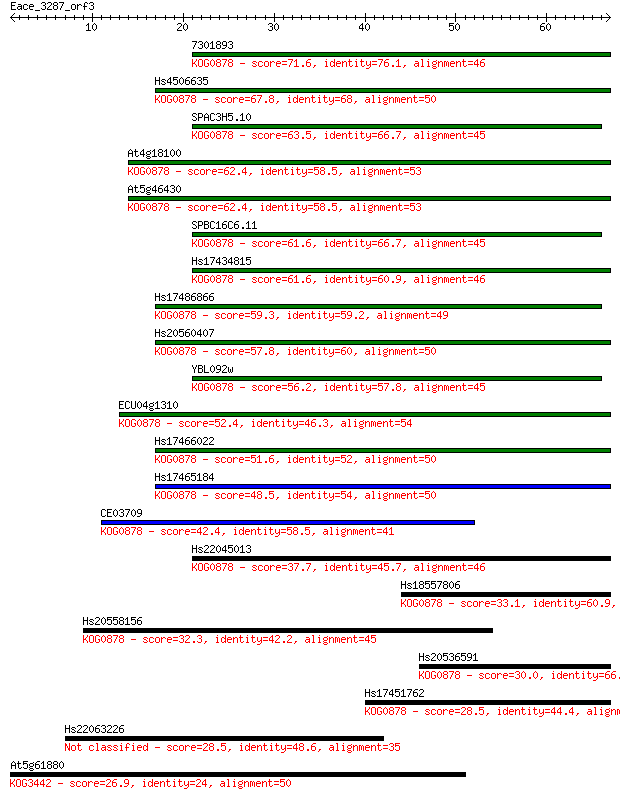
7301893 71.6 3e-13
Hs4506635 67.8 4e-12
SPAC3H5.10 63.5 1e-10
At4g18100 62.4 2e-10
At5g46430 62.4 2e-10
SPBC16C6.11 61.6 4e-10
Hs17434815 61.6 4e-10
Hs17486866 59.3 2e-09
Hs20560407 57.8 5e-09
YBL092w 56.2 1e-08
ECU04g1310 52.4 2e-07
Hs17466022 51.6 3e-07
Hs17465184 48.5 3e-06
CE03709 42.4 2e-04
Hs22045013 37.7 0.005
Hs18557806 33.1 0.14
Hs20558156 32.3 0.22
Hs20536591 30.0 1.1
Hs17451762 28.5 2.8
Hs22063226 28.5 3.3
At5g61880 26.9 9.8
> 7301893
Length=134
Score = 71.6 bits (174), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 35/46 (76%), Positives = 37/46 (80%), Gaps = 0/46 (0%)
Query 21 IVKKRTKRFNRFQSDRYKRLSPSWRKPKGIDCRVRRKFKGTNLMPN 66
IVKKRTK F R QSDRY +LS WRKPKGID RVRR+FKG LMPN
Sbjct 11 IVKKRTKHFIRHQSDRYAKLSHKWRKPKGIDNRVRRRFKGQYLMPN 56
> Hs4506635
Length=135
Score = 67.8 bits (164), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 34/50 (68%), Positives = 40/50 (80%), Gaps = 0/50 (0%)
Query 17 VKRTIVKKRTKRFNRFQSDRYKRLSPSWRKPKGIDCRVRRKFKGTNLMPN 66
VK IVKKRTK+F R QSDRY ++ +WRKP+GID RVRR+FKG LMPN
Sbjct 8 VKPKIVKKRTKKFIRHQSDRYVKIKRNWRKPRGIDNRVRRRFKGQILMPN 57
> SPAC3H5.10
Length=127
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 30/45 (66%), Positives = 36/45 (80%), Gaps = 0/45 (0%)
Query 21 IVKKRTKRFNRFQSDRYKRLSPSWRKPKGIDCRVRRKFKGTNLMP 65
I+KKRTK F R QSDR+KR+ SWRKP+GID VRR+F+GT MP
Sbjct 6 IIKKRTKPFKRHQSDRFKRVGESWRKPRGIDSCVRRRFRGTISMP 50
> At4g18100
Length=133
Score = 62.4 bits (150), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 31/53 (58%), Positives = 38/53 (71%), Gaps = 0/53 (0%)
Query 14 VSTVKRTIVKKRTKRFNRFQSDRYKRLSPSWRKPKGIDCRVRRKFKGTNLMPN 66
V + + +VKKR+ +F R QSDR + SWR+PKGID RVRRKFKG LMPN
Sbjct 3 VPLLTKKVVKKRSAKFIRPQSDRRITVKESWRRPKGIDSRVRRKFKGVTLMPN 55
> At5g46430
Length=133
Score = 62.4 bits (150), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 31/53 (58%), Positives = 38/53 (71%), Gaps = 0/53 (0%)
Query 14 VSTVKRTIVKKRTKRFNRFQSDRYKRLSPSWRKPKGIDCRVRRKFKGTNLMPN 66
V + + +VKKR+ +F R QSDR + SWR+PKGID RVRRKFKG LMPN
Sbjct 3 VPLLTKKVVKKRSAKFIRPQSDRRITVKESWRRPKGIDSRVRRKFKGVTLMPN 55
> SPBC16C6.11
Length=127
Score = 61.6 bits (148), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 30/45 (66%), Positives = 35/45 (77%), Gaps = 0/45 (0%)
Query 21 IVKKRTKRFNRFQSDRYKRLSPSWRKPKGIDCRVRRKFKGTNLMP 65
IVKKRTK F R QSD +KR+ SWRKP+GID VRR+F+GT MP
Sbjct 6 IVKKRTKPFKRHQSDLFKRVGESWRKPRGIDSCVRRRFRGTISMP 50
> Hs17434815
Length=115
Score = 61.6 bits (148), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 28/46 (60%), Positives = 36/46 (78%), Gaps = 0/46 (0%)
Query 21 IVKKRTKRFNRFQSDRYKRLSPSWRKPKGIDCRVRRKFKGTNLMPN 66
I+KKRTK+F QSDRY ++ P+W+KP+G D RV R+FKG LMPN
Sbjct 12 ILKKRTKKFIWHQSDRYVKIKPNWQKPRGADNRVHRRFKGQILMPN 57
> Hs17486866
Length=156
Score = 59.3 bits (142), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 29/49 (59%), Positives = 37/49 (75%), Gaps = 0/49 (0%)
Query 17 VKRTIVKKRTKRFNRFQSDRYKRLSPSWRKPKGIDCRVRRKFKGTNLMP 65
VK IVKKRTK+F +QSD Y ++ P+W+KP+GID RV R+FKG L P
Sbjct 29 VKLKIVKKRTKKFILYQSDWYVKIKPNWQKPRGIDNRVCRRFKGQILKP 77
> Hs20560407
Length=96
Score = 57.8 bits (138), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 30/50 (60%), Positives = 37/50 (74%), Gaps = 0/50 (0%)
Query 17 VKRTIVKKRTKRFNRFQSDRYKRLSPSWRKPKGIDCRVRRKFKGTNLMPN 66
VK I+KKRTK+F R QSDR + +W+KP+GID RV R+FKG LMPN
Sbjct 8 VKPKIIKKRTKKFIRHQSDRCVTIKRNWQKPRGIDNRVGRRFKGQILMPN 57
> YBL092w
Length=130
Score = 56.2 bits (134), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 26/45 (57%), Positives = 32/45 (71%), Gaps = 0/45 (0%)
Query 21 IVKKRTKRFNRFQSDRYKRLSPSWRKPKGIDCRVRRKFKGTNLMP 65
IVKK TK+F R SDRY R++ +WRK KGID VRR+F+G P
Sbjct 9 IVKKHTKKFKRHHSDRYHRVAENWRKQKGIDSVVRRRFRGNISQP 53
> ECU04g1310
Length=139
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 25/54 (46%), Positives = 36/54 (66%), Gaps = 2/54 (3%)
Query 13 PVSTVKRTIVKKRTKRFNRFQSDRYKRLSPSWRKPKGIDCRVRRKFKGTNLMPN 66
P+ +K ++ K+F R SDRYKR+ PSWR+P GID +VR++ KG MP+
Sbjct 10 PLVEIKEAY--RKNKKFIRHHSDRYKRVKPSWRRPHGIDSKVRKRCKGEREMPS 61
> Hs17466022
Length=345
Score = 51.6 bits (122), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 26/50 (52%), Positives = 36/50 (72%), Gaps = 0/50 (0%)
Query 17 VKRTIVKKRTKRFNRFQSDRYKRLSPSWRKPKGIDCRVRRKFKGTNLMPN 66
+K IVKKRTK+F QSD ++ +W +P+GID +V+R+FKG LMPN
Sbjct 218 LKPKIVKKRTKKFIWHQSDGQVQVKCNWWRPRGIDDKVQRRFKGQMLMPN 267
> Hs17465184
Length=139
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 27/50 (54%), Positives = 32/50 (64%), Gaps = 0/50 (0%)
Query 17 VKRTIVKKRTKRFNRFQSDRYKRLSPSWRKPKGIDCRVRRKFKGTNLMPN 66
+K IVKKRTK+F QSDR ++ +W KP I RV R FKG LMPN
Sbjct 8 MKPKIVKKRTKKFIWHQSDRCVKIKCNWWKPSSIGNRVWRTFKGQILMPN 57
> CE03709
Length=134
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/41 (58%), Positives = 31/41 (75%), Gaps = 0/41 (0%)
Query 11 MAPVSTVKRTIVKKRTKRFNRFQSDRYKRLSPSWRKPKGID 51
M VS K +VKK+ +F R +SDRY+R++PSWRKPKGID
Sbjct 1 MVHVSGTKVKVVKKKLTKFKRHESDRYRRVAPSWRKPKGID 41
> Hs22045013
Length=112
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 21/46 (45%), Positives = 31/46 (67%), Gaps = 2/46 (4%)
Query 21 IVKKRTKRFNRFQSDRYKRLSPSWRKPKGIDCRVRRKFKGTNLMPN 66
I+KKR+K+F QS++Y ++ + KP+ D V R+FKG LMPN
Sbjct 12 IIKKRSKKFIHHQSEQYVKIKHNRWKPR--DNCVHRRFKGQILMPN 55
> Hs18557806
Length=222
Score = 33.1 bits (74), Expect = 0.14, Method: Composition-based stats.
Identities = 14/23 (60%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 44 WRKPKGIDCRVRRKFKGTNLMPN 66
W+KP+G D RV R+FK LMPN
Sbjct 133 WQKPRGSDNRVHRRFKLQILMPN 155
> Hs20558156
Length=235
Score = 32.3 bits (72), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 19/45 (42%), Positives = 28/45 (62%), Gaps = 2/45 (4%)
Query 9 STMAPVSTVKRTIVKKRTKRFNRFQSDRYKRLSPSWRKPKGIDCR 53
+T+ P+ VK IVK RTK+F QSD Y ++ W+KP+ + R
Sbjct 14 ATLRPL--VKPRIVKNRTKKFIWHQSDPYVKIKCKWQKPRASNTR 56
> Hs20536591
Length=248
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 14/21 (66%), Positives = 16/21 (76%), Gaps = 0/21 (0%)
Query 46 KPKGIDCRVRRKFKGTNLMPN 66
KP+GID RV R+ KG LMPN
Sbjct 7 KPRGIDNRVCRRCKGQILMPN 27
> Hs17451762
Length=112
Score = 28.5 bits (62), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 12/27 (44%), Positives = 17/27 (62%), Gaps = 0/27 (0%)
Query 40 LSPSWRKPKGIDCRVRRKFKGTNLMPN 66
L + P+GID R++R+FK LM N
Sbjct 8 LGELCQTPRGIDNRIQRRFKSQLLMSN 34
> Hs22063226
Length=419
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 17/39 (43%), Positives = 23/39 (58%), Gaps = 4/39 (10%)
Query 7 PASTM----APVSTVKRTIVKKRTKRFNRFQSDRYKRLS 41
P S+M AP S V VK RT++ N FQ+++ K LS
Sbjct 336 PCSSMPAKQAPPSCVSEGSVKGRTQKENLFQTNKLKSLS 374
> At5g61880
Length=525
Score = 26.9 bits (58), Expect = 9.8, Method: Composition-based stats.
Identities = 12/50 (24%), Positives = 27/50 (54%), Gaps = 0/50 (0%)
Query 1 RKERHKPASTMAPVSTVKRTIVKKRTKRFNRFQSDRYKRLSPSWRKPKGI 50
R+++ +P S M P ++++ K++T +NR ++ LS +P +
Sbjct 353 RRKQQEPESPMGPKELIRQSDTKQKTLVYNRRRTRSQALLSSQTARPDAL 402
Lambda K H
0.321 0.132 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1163608362
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40