bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_3180_orf1
Length=153
Score E
Sequences producing significant alignments: (Bits) Value
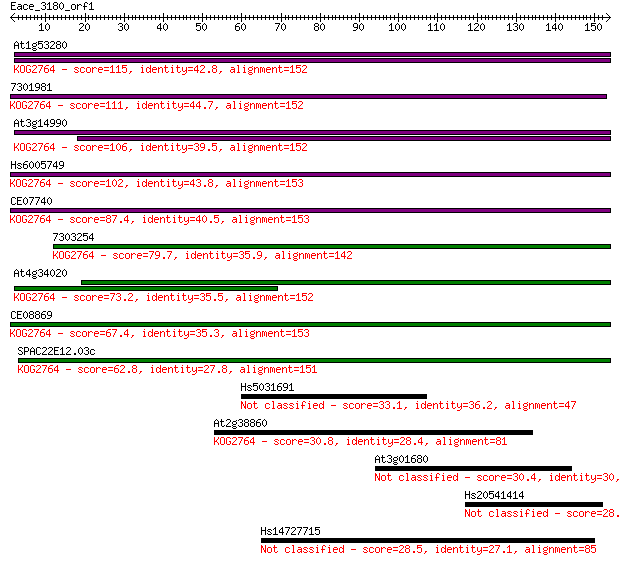
At1g53280 115 3e-26
7301981 111 7e-25
At3g14990 106 1e-23
Hs6005749 102 2e-22
CE07740 87.4 9e-18
7303254 79.7 2e-15
At4g34020 73.2 2e-13
CE08869 67.4 1e-11
SPAC22E12.03c 62.8 3e-10
Hs5031691 33.1 0.21
At2g38860 30.8 1.1
At3g01680 30.4 1.6
Hs20541414 28.9 3.7
Hs14727715 28.5 5.7
> At1g53280
Length=438
Score = 115 bits (288), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 65/154 (42%), Positives = 100/154 (64%), Gaps = 3/154 (1%)
Query 2 LVILANGAEDIEFASPVDVLRRAGVKVVVASLGEGLIVKTAQGLRIEGDMPLEDVSPDAH 61
LV +A+G+E++E + +DVL+RA VVVA+LG L V ++ +++ D+ L++ ++
Sbjct 261 LVPIADGSEEMEAVAIIDVLKRAKANVVVAALGNSLEVVASRKVKLVADVLLDEAEKNS- 319
Query 62 YDVIVLPGGLKGAENCRDSAHLASLLKQQHAAGRWIAAICASPALVLTHHGLLENINSVA 121
YD+IVLPGGL GAE S L ++LK+Q + + AICASPALV HGLL+ + A
Sbjct 320 YDLIVLPGGLGGAEAFASSEKLVNMLKKQAESNKPYGAICASPALVFEPHGLLKGKKATA 379
Query 122 YPSFMSHIKHKG--EGRVCVDKHYITSIGPGSAM 153
+P+ S + + E RV VD + ITS GPG+++
Sbjct 380 FPAMCSKLTDQSHIEHRVLVDGNLITSRGPGTSL 413
Score = 105 bits (262), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 60/155 (38%), Positives = 86/155 (55%), Gaps = 4/155 (2%)
Query 2 LVILANGAEDIEFASPVDVLRRAGVKVVVASLGEGLIVKTAQGLRIEGDMPLEDVSPDAH 61
L+ +A+G E E +DVLRR G V VAS+ + V G+++ D L D++ D+
Sbjct 56 LIPVAHGTEPFEAVVMIDVLRRGGADVTVASVENQVGVDACHGIKMVADTLLSDIT-DSV 114
Query 62 YDVIVLPGGLKGAENCRDSAHLASLLKQQHAAGRWIAAICASPALVLTHHGLLENINSVA 121
+D+I+LPGGL G E ++ L ++K+Q GR AAIC +PAL GLLE +
Sbjct 115 FDLIMLPGGLPGGETLKNCKPLEKMVKKQDTDGRLNAAICCAPALAFGTWGLLEGKKATC 174
Query 122 YPSFMSHIKHKG---EGRVCVDKHYITSIGPGSAM 153
YP FM + E RV +D +TS GPG+ M
Sbjct 175 YPVFMEKLAACATAVESRVEIDGKIVTSRGPGTTM 209
> 7301981
Length=187
Score = 111 bits (277), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 68/156 (43%), Positives = 95/156 (60%), Gaps = 6/156 (3%)
Query 1 ALVILANGAEDIEFASPVDVLRRAGVKVVVASLGEGLIVKTAQGLRIEGDMPLEDVSPDA 60
ALVILA GAE++EF DVLRRAG+KV VA L G VK ++ ++I D L V+ D
Sbjct 5 ALVILAPGAEEMEFIIAADVLRRAGIKVTVAGLNGGEAVKCSRDVQILPDTSLAQVASD- 63
Query 61 HYDVIVLPGGLKGAENCRDSAHLASLLKQQHAAGRWIAAICASPALVLTHHGLLENINSV 120
+DV+VLPGGL G+ +S+ + LL+ Q + G IAAICA+P VL HG+ +
Sbjct 64 KFDVVVLPGGLGGSNAMGESSLVGDLLRSQESGGGLIAAICAAPT-VLAKHGVASGKSLT 122
Query 121 AYPS----FMSHIKHKGEGRVCVDKHYITSIGPGSA 152
+YPS +++ + + V D + ITS GPG+A
Sbjct 123 SYPSMKPQLVNNYSYVDDKTVVKDGNLITSRGPGTA 158
> At3g14990
Length=392
Score = 106 bits (265), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 60/156 (38%), Positives = 90/156 (57%), Gaps = 5/156 (3%)
Query 2 LVILANGAEDIEFASPVDVLRRAGVKVVVASLGEGLIVKTAQGLRIEGDMPLEDVSPDAH 61
L+ +A+G E +E + + VLRR G V VAS+ + V G+++ D L D++ D+
Sbjct 9 LIPIAHGTEPLEAVAMITVLRRGGADVTVASVETQVGVDACHGIKMVADTLLSDIT-DSV 67
Query 62 YDVIVLPGGLKGAENCRDSAHLASLLKQQHAAGRWIAAICASPALVLTHHGLLENINSVA 121
+D+IVLPGGL G E ++ L +++K+Q + GR AAIC +PAL L GLLE +
Sbjct 68 FDLIVLPGGLPGGETLKNCKSLENMVKKQDSDGRLNAAICCAPALALGTWGLLEGKKATG 127
Query 122 YPSFMSHIKHKG----EGRVCVDKHYITSIGPGSAM 153
YP FM + E RV +D +TS GPG+ +
Sbjct 128 YPVFMEKLAATCATAVESRVQIDGRIVTSRGPGTTI 163
Score = 92.8 bits (229), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 55/142 (38%), Positives = 85/142 (59%), Gaps = 11/142 (7%)
Query 18 VDVLRRAGVKVVVASLGEGLIVKTAQGLRIEGDMPLEDVSPDAHYDVIVLPGGLKGAENC 77
VD+LRRA VV+A++G L V+ ++ ++ ++ L++V+ + +D+IVLPGGL GA+
Sbjct 231 VDILRRAKANVVIAAVGNSLEVEGSRKAKLVAEVLLDEVA-EKSFDLIVLPGGLNGAQRF 289
Query 78 RDSAHLASLLKQQHAAGRWIAAICASPALVLTHHGLLENINSVAYP------SFMSHIKH 131
L ++L++Q A + ICASPA V +GLL+ + +P S SHI+H
Sbjct 290 ASCEKLVNMLRKQAEANKPYGGICASPAYVFEPNGLLKGKKATTHPVVSDKLSDKSHIEH 349
Query 132 KGEGRVCVDKHYITSIGPGSAM 153
RV VD + ITS PG+AM
Sbjct 350 ----RVVVDGNVITSRAPGTAM 367
> Hs6005749
Length=189
Score = 102 bits (255), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 67/158 (42%), Positives = 90/158 (56%), Gaps = 6/158 (3%)
Query 1 ALVILANGAEDIEFASPVDVLRRAGVKVVVASLGEGLIVKTAQGLRIEGDMPLEDVSPDA 60
ALVILA GAE++E PVDV+RRAG+KV VA L V+ ++ + I D LED +
Sbjct 6 ALVILAKGAEEMETVIPVDVMRRAGIKVTVAGLAGKDPVQCSRDVVICPDASLEDAKKEG 65
Query 61 HYDVIVLPGGLKGAENCRDSAHLASLLKQQHAAGRWIAAICASPALVLTHH---GLLENI 117
YDV+VLPGG GA+N +SA + +LK+Q IAAICA P +L H G
Sbjct 66 PYDVVVLPGGNLGAQNLSESAAVKEILKEQENRKGLIAAICAGPTALLAHEIGCGSKVTT 125
Query 118 NSVAYPSFMS--HIKHKGEGRVCVDKHYITSIGPGSAM 153
+ +A M+ H + E RV D +TS GPG++
Sbjct 126 HPLAKDKMMNGGHYTY-SENRVEKDGLILTSRGPGTSF 162
> CE07740
Length=187
Score = 87.4 bits (215), Expect = 9e-18, Method: Compositional matrix adjust.
Identities = 62/158 (39%), Positives = 87/158 (55%), Gaps = 8/158 (5%)
Query 1 ALVILA-NGAEDIEFASPVDVLRRAGVKVVVASLGEGLIVKTAQGLRIEGDMPLEDVSPD 59
AL+ILA GAE++E DVL R ++VV A L VK A+G I D+ LEDV +
Sbjct 6 ALIILAAEGAEEMEVIITGDVLARGEIRVVYAGLDGAEPVKCARGAHIVPDVKLEDVETE 65
Query 60 AHYDVIVLPGGLKGAENCRDSAHLASLLKQQHAAGRWIAAICASPALVLTHHGLLENINS 119
+D+++LPGG G+ +S + +LK Q +G I AICA+P +L+H E + S
Sbjct 66 -KFDIVILPGGQPGSNTLAESLLVRDVLKSQVESGGLIGAICAAPIALLSHGVKAELVTS 124
Query 120 VAYPSFMSHIKHKG----EGRVCVDKHYITSIGPGSAM 153
+PS ++ G E RV V ITS GPG+A
Sbjct 125 --HPSVKEKLEKGGYKYSEDRVVVSGKIITSRGPGTAF 160
> 7303254
Length=174
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 51/146 (34%), Positives = 77/146 (52%), Gaps = 5/146 (3%)
Query 12 IEFASPVDVLRRAGVKVVVASLGEGLIVKTAQGLRIEGDMPLEDVSPDAHYDVIVLPGGL 71
+EF DVLRR + V VA L + VK ++ + I D LE+ YDV+VLPGGL
Sbjct 1 MEFTISADVLRRGKILVTVAGLHDCEPVKCSRSVVIVPDTSLEEAVTRGDYDVVVLPGGL 60
Query 72 KGAENCRDSAHLASLLKQQHAAGRWIAAICASPALVLTHHGLLENINSVAYPSFMSHIK- 130
G + +S+ + +L+ Q + G IAAICA+P L HG+ + + ++P +K
Sbjct 61 AGNKALMNSSAVGDVLRCQESKGGLIAAICAAPT-ALAKHGIGKGKSITSHPDMKPQLKE 119
Query 131 ---HKGEGRVCVDKHYITSIGPGSAM 153
+ + V D + ITS GPG+
Sbjct 120 LYCYIDDKTVVQDGNIITSRGPGTTF 145
> At4g34020
Length=334
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 47/135 (34%), Positives = 70/135 (51%), Gaps = 9/135 (6%)
Query 19 DVLRRAGVKVVVASLGEGLIVKTAQGLRIEGDMPLEDVSPDAHYDVIVLPGGLKGAENCR 78
DVLRRAG V VAS+ + L V+ + G R+ D+ + + D YD++ LPGG+ GA R
Sbjct 116 DVLRRAGADVTVASVEQKLEVEGSSGTRLLADVLISKCA-DQVYDLVALPGGMPGAVRLR 174
Query 79 DSAHLASLLKQQHAAGRWIAAICASPALVLTHHGLLENINSVAYPSFMSHIKHKGEGRVC 138
D L ++K+Q R AI +PA+ L GLL + + ++I+ GE
Sbjct 175 DCEILEKIMKRQAEDKRLYGAISMAPAITLLPWGLLTRKRLPTFWAVKTNIQISGE---- 230
Query 139 VDKHYITSIGPGSAM 153
TS GPG++
Sbjct 231 ----LTTSRGPGTSF 241
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 42/67 (62%), Gaps = 1/67 (1%)
Query 2 LVILANGAEDIEFASPVDVLRRAGVKVVVASLGEGLIVKTAQGLRIEGDMPLEDVSPDAH 61
L+ +ANG+E +E S DVLRRA V V V+S+ L + QG +I D + + + ++
Sbjct 266 LIPVANGSEAVELVSIADVLRRAKVDVTVSSVERSLRITAFQGTKIITDKLIGEAA-ESS 324
Query 62 YDVIVLP 68
YD+I+LP
Sbjct 325 YDLIILP 331
> CE08869
Length=186
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 54/156 (34%), Positives = 80/156 (51%), Gaps = 6/156 (3%)
Query 1 ALVILA-NGAEDIEFASPVDVLRRAGVKVVVASLGEGLIVKTAQGLRIEGDMPLEDVSPD 59
AL++L AE+IE DVL R G++V+ A VK A+G RI D+ L+DV +
Sbjct 7 ALILLPPEDAEEIEVIVTGDVLVRGGLQVLYAG-SSTEPVKCAKGARIVPDVALKDVK-N 64
Query 60 AHYDVIVLPGGLKGAENCRDSAHLASLLKQQHAAGRWIAAICASPALVLTHHGLLENINS 119
+D+I++PGG G + + LLK Q +G I AICA P ++L H + E +
Sbjct 65 KTFDIIIIPGG-PGCSKLAECPVIGELLKTQVKSGGLIGAICAGPTVLLAHGIVAERVTC 123
Query 120 -VAYPSFMSHIKHKG-EGRVCVDKHYITSIGPGSAM 153
M+ +K + V + ITS GPG+A
Sbjct 124 HYTVKDKMTEGGYKYLDDNVVISDRVITSKGPGTAF 159
> SPAC22E12.03c
Length=191
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 42/163 (25%), Positives = 83/163 (50%), Gaps = 15/163 (9%)
Query 3 VILANGAEDIEFASPVDVLRRAGVKVVVASLGEG--LIVKTAQGLRIEGDMPLEDVSPDA 60
+ +A+G ++IEF++P + +RA + + +GE +VK ++ + + + +++ P A
Sbjct 6 LFVADGTDEIEFSAPWGIFKRAEIPIDSVYVGENKDRLVKMSRDVEMYANRSYKEI-PSA 64
Query 61 -----HYDVIVLPGGLKGAENCRDSAHLASLLKQQHAA-GRWIAAICASPALVLTHHGLL 114
YD+ ++PGG GA+ + + ++K+ + +WI ICA T L
Sbjct 65 DDFAKQYDIAIIPGGGLGAKTLSTTPFVQQVVKEFYKKPNKWIGMICAGTLTAKTSG--L 122
Query 115 ENINSVAYPSFMSHIKHKG----EGRVCVDKHYITSIGPGSAM 153
N +PS ++ G + V ++++ ITS GPG+AM
Sbjct 123 PNKQITGHPSVRGQLEEGGYKYLDQPVVLEENLITSQGPGTAM 165
> Hs5031691
Length=268
Score = 33.1 bits (74), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 28/57 (49%), Gaps = 10/57 (17%)
Query 60 AHYDVIVLPGGLKGAEN----------CRDSAHLASLLKQQHAAGRWIAAICASPAL 106
A++D + PGG A+N C+ + + +LK+ H AG+ I C +P L
Sbjct 126 ANHDAAIFPGGFGAAKNLSTFAVDGKDCKVNKEVERVLKEFHQAGKPIGLCCIAPVL 182
> At2g38860
Length=398
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 23/86 (26%), Positives = 39/86 (45%), Gaps = 12/86 (13%)
Query 53 LEDVSPDAHYDVIVLPGG-----LKGAENCRDSAHLASLLKQQHAAGRWIAAICASPALV 107
+DV+P+ +YDVI++PGG L E C D L+ + + + I C S ++
Sbjct 76 FDDVTPE-NYDVIIIPGGRFTELLSADEKCVD------LVARFAESKKLIFTSCHSQVML 128
Query 108 LTHHGLLENINSVAYPSFMSHIKHKG 133
+ L + A+ S I+ G
Sbjct 129 MAAGILAGGVKCTAFESIKPLIELSG 154
> At3g01680
Length=740
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 15/50 (30%), Positives = 25/50 (50%), Gaps = 1/50 (2%)
Query 94 GRWIAAICASPALVLTHHGLLENINSVAYPSFMSHIKHKGEGRVCVDKHY 143
G W A + P +V+ HG +E SV ++ +H+ KG + D H+
Sbjct 632 GGW-ALLSKGPEIVMIAHGAIERTMSVYDRTWKTHVPTKGYTKAMSDHHH 680
> Hs20541414
Length=428
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 13/35 (37%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 117 INSVAYPSFMSHIKHKGEGRVCVDKHYITSIGPGS 151
S+ Y ++ +KHK + K YI S+GPGS
Sbjct 343 TQSLGYGAYTQRLKHKAYTQSRGHKAYIQSLGPGS 377
> Hs14727715
Length=1294
Score = 28.5 bits (62), Expect = 5.7, Method: Composition-based stats.
Identities = 23/86 (26%), Positives = 37/86 (43%), Gaps = 5/86 (5%)
Query 65 IVLPGGLKGAENCRDSAHLASLLKQQHAAGRWIAAICASPALVLTHHGL-LENINSVAYP 123
I+L G A + + A + L QQ+++G W +CA L H E + Y
Sbjct 428 ILLEGSQLDASDWLNPAQVV-LFSQQNSSGPWAMDLCARRLLDPCEHQCDPETGECLCYE 486
Query 124 SFMSHIKHKGEGRVCVDKHYITSIGP 149
+M HK +C+ + T+ GP
Sbjct 487 GYMKDPVHK---HLCIRNEWGTNQGP 509
Lambda K H
0.319 0.136 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1961355924
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40