bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_3083_orf3
Length=165
Score E
Sequences producing significant alignments: (Bits) Value
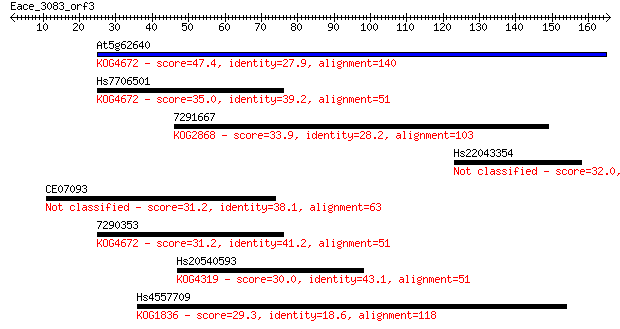
At5g62640 47.4 1e-05
Hs7706501 35.0 0.070
7291667 33.9 0.15
Hs22043354 32.0 0.51
CE07093 31.2 0.89
7290353 31.2 1.1
Hs20540593 30.0 2.5
Hs4557709 29.3 3.3
> At5g62640
Length=520
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 39/140 (27%), Positives = 71/140 (50%), Gaps = 11/140 (7%)
Query 25 KAYNPSTAHRRKERDAEKKKNKAERKITREQALLKKNPVVIKEELDKLEEMARSGQTMTE 84
K NP+ A+R++ R E K+NK ER+ RE +LKK+P IK+++ KL +M+++ + +
Sbjct 8 KVMNPTDAYRKQIRKREIKRNKKERQKVREVGILKKDPEQIKDQIRKL-DMSKAEGALDK 66
Query 85 GRTRRLQKLQQLWAQVVSSVEAGRRVRTADVSSVASVDPRSSSSSSTVNARCLQQRLSNG 144
R + ++L+ VV ++ + D + +++S + + QR G
Sbjct 67 ARKHKKRQLEDTLKMVV------KKRKEYDEKKKE----QGEATTSVMFSHLPPQRRLTG 116
Query 145 ERVPDEDERRYYCPELCNTG 164
E ++ YY P L TG
Sbjct 117 EEDLKPEDSVYYHPTLNPTG 136
> Hs7706501
Length=641
Score = 35.0 bits (79), Expect = 0.070, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 30/51 (58%), Gaps = 0/51 (0%)
Query 25 KAYNPSTAHRRKERDAEKKKNKAERKITREQALLKKNPVVIKEELDKLEEM 75
K NP+ R++ R E KKNK +R + R L K+P I +++KL+EM
Sbjct 13 KFMNPTDQARKEARKRELKKNKKQRMMVRAAVLKMKDPKQIIRDMEKLDEM 63
> 7291667
Length=372
Score = 33.9 bits (76), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 29/112 (25%), Positives = 57/112 (50%), Gaps = 13/112 (11%)
Query 46 KAERKITREQALLKKNPVVIKEELDKLEEMARSGQTMT--------EGRTRRLQKLQQLW 97
++ ++ T E + P+ + ++L+K + A G+ + E R QK+QQL
Sbjct 193 ESAKQATAESLFHRVQPLSV-DQLEKQQRAATPGEDLLPPAGRDNHESRLSPFQKMQQLN 251
Query 98 AQVVSSVEAGRRVRTADVSSVASVDPRSSSSSSTVNA-RCLQQRLSNGERVP 148
Q++S ++ G+ TA +VA P SS+ + +A +CL++ L+ + P
Sbjct 252 IQLLSDLKIGKSYATAQSPTVA---PSSSALMANDDAGKCLRRLLAGDKPGP 300
> Hs22043354
Length=619
Score = 32.0 bits (71), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 123 PRSSSSSSTVNARCLQQRLSNGERVPDEDERRYYC 157
P + SSS++VN+ + L NGE V D R Y+C
Sbjct 215 PTTVSSSASVNSCAVNPCLHNGECVADNTSRGYHC 249
> CE07093
Length=361
Score = 31.2 bits (69), Expect = 0.89, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 35/73 (47%), Gaps = 12/73 (16%)
Query 11 MFNPSKKFIGVETLKA----------YNPSTAHRRKERDAEKKKNKAERKITREQALLKK 60
M NP G++ L A YN S A R+ R KK K +K++ +Q +L+K
Sbjct 109 MSNPLILMTGIDRLIACKSPVLKKTKYNHSDA--RRLRCQHKKSTKLTKKMSPKQLILRK 166
Query 61 NPVVIKEELDKLE 73
NP+ I+ LE
Sbjct 167 NPISIRRYRALLE 179
> 7290353
Length=552
Score = 31.2 bits (69), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 21/51 (41%), Positives = 32/51 (62%), Gaps = 0/51 (0%)
Query 25 KAYNPSTAHRRKERDAEKKKNKAERKITREQALLKKNPVVIKEELDKLEEM 75
K NP+ R++ R E KKNK +R++ R L K+P I EE++K++EM
Sbjct 13 KYMNPTDQARKEARKKELKKNKKQRQMVRAAVLKNKDPSQILEEMEKIDEM 63
> Hs20540593
Length=97
Score = 30.0 bits (66), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 22/58 (37%), Positives = 29/58 (50%), Gaps = 7/58 (12%)
Query 47 AERKITREQALLKKNPVVIKEE-----LDK--LEEMARSGQTMTEGRTRRLQKLQQLW 97
AER + R QAL + P + EE LD L + G ++GRTRR Q L+ W
Sbjct 5 AERALPRLQALARPPPPISYEEELYDCLDYYYLRDFPACGAGRSKGRTRREQALRTNW 62
> Hs4557709
Length=3110
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 22/118 (18%), Positives = 49/118 (41%), Gaps = 2/118 (1%)
Query 36 KERDAEKKKNKAERKITREQALLKKNPVVIKEELDKLEEMARSGQTMTEGRTRRLQKLQQ 95
+ ++ E +K AE ++ +ALLKK + E + EEM + + ++
Sbjct 1729 RRKNLETQKEIAEDELVAAEALLKKVKKLFGESRGENEEMEKDLREKLADYKNKVDDAWD 1788
Query 96 LWAQVVSSVEAGRRVRTADVSSVASVDPRSSSSSSTVNARCLQQRLSNGERVPDEDER 153
L + + R+ + ++ +++ + + S R ++ L G + DE R
Sbjct 1789 LLREATDKIREANRLFAVNQKNMTALEKKKEAVES--GKRQIENTLKEGNDILDEANR 1844
Lambda K H
0.314 0.127 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2353551590
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40