bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2990_orf1
Length=163
Score E
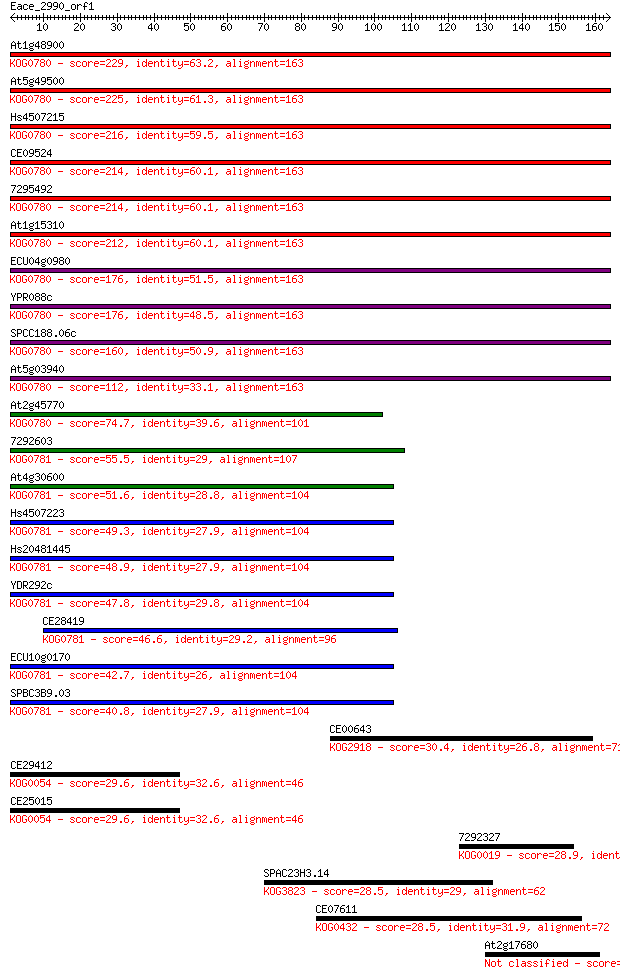
Sequences producing significant alignments: (Bits) Value
At1g48900 229 2e-60
At5g49500 225 4e-59
Hs4507215 216 1e-56
CE09524 214 5e-56
7295492 214 8e-56
At1g15310 212 3e-55
ECU04g0980 176 1e-44
YPR088c 176 2e-44
SPCC188.06c 160 1e-39
At5g03940 112 2e-25
At2g45770 74.7 8e-14
7292603 55.5 5e-08
At4g30600 51.6 7e-07
Hs4507223 49.3 4e-06
Hs20481445 48.9 4e-06
YDR292c 47.8 9e-06
CE28419 46.6 2e-05
ECU10g0170 42.7 3e-04
SPBC3B9.03 40.8 0.001
CE00643 30.4 1.6
CE29412 29.6 2.4
CE25015 29.6 2.4
7292327 28.9 4.6
SPAC23H3.14 28.5 5.3
CE07611 28.5 6.7
At2g17680 28.5 6.8
> At1g48900
Length=522
Score = 229 bits (584), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 103/163 (63%), Positives = 137/163 (84%), Gaps = 0/163 (0%)
Query 1 SGRHRQEAALFEEMQQVADAVNPDDIVFVVDSHIGQACYDQAAAFAASVSVGSVIITKLD 60
SGRH+QEA+LFEEM+QVA+A PD ++FV+DS IGQA +DQA AF SV+VG+VIITK+D
Sbjct 219 SGRHKQEASLFEEMRQVAEATKPDLVIFVMDSSIGQAAFDQAQAFKQSVAVGAVIITKMD 278
Query 61 GYAKGGGALSAVAATGAPIIFLGSGEHFDDFDPFDANSFVSRLLGLGDMGGLVNTLKEVI 120
G+AKGGGALSAVAAT +P+IF+G+GEH D+F+ FD FVSRLLG+GD G V+ L+EV+
Sbjct 279 GHAKGGGALSAVAATKSPVIFIGTGEHMDEFEVFDVKPFVSRLLGMGDWSGFVDKLQEVV 338
Query 121 SAEKQQEMIERISKGKFTLQDMYDQFQNVLKMGPISKVMSMIP 163
++Q E++E++S+G FTL+ MYDQFQN+L MGP+ +V SM+P
Sbjct 339 PKDQQPELLEKLSQGNFTLRIMYDQFQNILNMGPLKEVFSMLP 381
> At5g49500
Length=497
Score = 225 bits (573), Expect = 4e-59, Method: Compositional matrix adjust.
Identities = 100/163 (61%), Positives = 135/163 (82%), Gaps = 0/163 (0%)
Query 1 SGRHRQEAALFEEMQQVADAVNPDDIVFVVDSHIGQACYDQAAAFAASVSVGSVIITKLD 60
SGRH+Q+A+LFEEM+Q+++A PD ++FV+DS IGQ ++QA AF +V+VG+VIITK+D
Sbjct 194 SGRHKQQASLFEEMRQISEATKPDLVIFVMDSSIGQTAFEQARAFKQTVAVGAVIITKMD 253
Query 61 GYAKGGGALSAVAATGAPIIFLGSGEHFDDFDPFDANSFVSRLLGLGDMGGLVNTLKEVI 120
G+AKGGG LSAVAAT +P+IF+G+GEH D+F+ FDA FVSRLLG GDM G VN L+EV+
Sbjct 254 GHAKGGGTLSAVAATKSPVIFIGTGEHMDEFEVFDAKPFVSRLLGNGDMSGFVNKLQEVV 313
Query 121 SAEKQQEMIERISKGKFTLQDMYDQFQNVLKMGPISKVMSMIP 163
++Q E++E +S GKFTL+ MYDQFQN+L MGP+ +V SM+P
Sbjct 314 PKDQQPELLEMLSHGKFTLRIMYDQFQNMLNMGPLKEVFSMLP 356
> Hs4507215
Length=504
Score = 216 bits (550), Expect = 1e-56, Method: Compositional matrix adjust.
Identities = 97/163 (59%), Positives = 132/163 (80%), Gaps = 1/163 (0%)
Query 1 SGRHRQEAALFEEMQQVADAVNPDDIVFVVDSHIGQACYDQAAAFAASVSVGSVIITKLD 60
SGRH+QE +LFEEM QVA+A+ PD+IV+V+D+ IGQAC QA AF V V SVI+TKLD
Sbjct 192 SGRHKQEDSLFEEMLQVANAIQPDNIVYVMDASIGQACEAQAKAFKDKVDVASVIVTKLD 251
Query 61 GYAKGGGALSAVAATGAPIIFLGSGEHFDDFDPFDANSFVSRLLGLGDMGGLVNTLKEVI 120
G+AKGGGALSAVAAT +PIIF+G+GEH DDF+PF F+S+LLG+GD+ GL++ + E +
Sbjct 252 GHAKGGGALSAVAATKSPIIFIGTGEHIDDFEPFKTQPFISKLLGMGDIEGLIDKVNE-L 310
Query 121 SAEKQQEMIERISKGKFTLQDMYDQFQNVLKMGPISKVMSMIP 163
+ + +IE++ G+FTL+DMY+QFQN++KMGP S+++ MIP
Sbjct 311 KLDDNEALIEKLKHGQFTLRDMYEQFQNIMKMGPFSQILGMIP 353
> CE09524
Length=496
Score = 214 bits (545), Expect = 5e-56, Method: Compositional matrix adjust.
Identities = 98/163 (60%), Positives = 133/163 (81%), Gaps = 1/163 (0%)
Query 1 SGRHRQEAALFEEMQQVADAVNPDDIVFVVDSHIGQACYDQAAAFAASVSVGSVIITKLD 60
SGRH+QEA+LFEEM QV++AV PD++VFV+D+ IGQAC QA AF+ +V V SVIITKLD
Sbjct 192 SGRHKQEASLFEEMLQVSNAVTPDNVVFVMDASIGQACEAQARAFSQTVDVASVIITKLD 251
Query 61 GYAKGGGALSAVAATGAPIIFLGSGEHFDDFDPFDANSFVSRLLGLGDMGGLVNTLKEVI 120
+AKGGGALSAVA T +P+IF+G+GEH DDF+ F SFV +LLG+GD+ GLV+ + + I
Sbjct 252 SHAKGGGALSAVAVTKSPVIFIGTGEHIDDFEIFKPKSFVQKLLGMGDIAGLVDMVND-I 310
Query 121 SAEKQQEMIERISKGKFTLQDMYDQFQNVLKMGPISKVMSMIP 163
+ +E++ R+ +G+FTL+DMY+QFQN++KMGP S++M MIP
Sbjct 311 GIQDNKELVGRLKQGQFTLRDMYEQFQNIMKMGPFSQIMGMIP 353
> 7295492
Length=508
Score = 214 bits (544), Expect = 8e-56, Method: Compositional matrix adjust.
Identities = 98/163 (60%), Positives = 131/163 (80%), Gaps = 1/163 (0%)
Query 1 SGRHRQEAALFEEMQQVADAVNPDDIVFVVDSHIGQACYDQAAAFAASVSVGSVIITKLD 60
SGRH+QE +LFEEM V++AV+PD+I+FV+D+ IGQAC QA AF V +GSVIITKLD
Sbjct 192 SGRHKQEESLFEEMLAVSNAVSPDNIIFVMDATIGQACEAQAKAFKDKVDIGSVIITKLD 251
Query 61 GYAKGGGALSAVAATGAPIIFLGSGEHFDDFDPFDANSFVSRLLGLGDMGGLVNTLKEVI 120
G+AKGGGALSAVAAT +PIIF+G+GEH DD +PF FVS+LLG+GD+ GL++ + E +
Sbjct 252 GHAKGGGALSAVAATQSPIIFIGTGEHIDDLEPFKTKPFVSKLLGMGDIEGLIDKVNE-L 310
Query 121 SAEKQQEMIERISKGKFTLQDMYDQFQNVLKMGPISKVMSMIP 163
+ E++E+I G FT++DMY+QFQN++KMGP S+ M+MIP
Sbjct 311 KLDGNDELLEKIKHGHFTIRDMYEQFQNIMKMGPFSQFMNMIP 353
> At1g15310
Length=479
Score = 212 bits (539), Expect = 3e-55, Method: Compositional matrix adjust.
Identities = 98/163 (60%), Positives = 133/163 (81%), Gaps = 1/163 (0%)
Query 1 SGRHRQEAALFEEMQQVADAVNPDDIVFVVDSHIGQACYDQAAAFAASVSVGSVIITKLD 60
SGRH+Q A+LFEEM+QVA+A PD ++FV+DS IGQA ++QA AF +VSVG+VIITK+D
Sbjct 192 SGRHKQAASLFEEMRQVAEATEPDLVIFVMDSSIGQAAFEQAEAFKETVSVGAVIITKMD 251
Query 61 GYAKGGGALSAVAATGAPIIFLGSGEHFDDFDPFDANSFVSRLLGLGDMGGLVNTLKEVI 120
G+AKGGGALSAVAAT +P+IF+G+GEH D+F+ FD FVSRLLG GD GLV+ L+EV+
Sbjct 252 GHAKGGGALSAVAATKSPVIFIGTGEHMDEFEVFDVKPFVSRLLGKGDWSGLVDKLQEVV 311
Query 121 SAEKQQEMIERISKGKFTLQDMYDQFQNVLKMGPISKVMSMIP 163
+ Q E++E +S+G FTL+ MYDQFQ L++ P++++ SM+P
Sbjct 312 PKDLQNELVENLSQGNFTLRSMYDQFQCSLRI-PLNQLFSMLP 353
> ECU04g0980
Length=466
Score = 176 bits (447), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 84/163 (51%), Positives = 117/163 (71%), Gaps = 2/163 (1%)
Query 1 SGRHRQEAALFEEMQQVADAVNPDDIVFVVDSHIGQACYDQAAAFAASVSVGSVIITKLD 60
SGRH QE LF EM+ + ++P IVFV+D+ IGQ+ DQA F +V VGS+I+TK+D
Sbjct 189 SGRHTQETELFTEMKDIIREISPSSIVFVMDAGIGQSAEDQAMGFKRAVDVGSIILTKID 248
Query 61 GYAKGGGALSAVAATGAPIIFLGSGEHFDDFDPFDANSFVSRLLGLGDMGGLVNTLKEVI 120
G K GGA+S+VAAT PI F+G+GE DD + FDA FVSR+LG+GD+ GL+ + +
Sbjct 249 GTTKAGGAISSVAATKCPIEFVGTGEGMDDLEAFDARRFVSRMLGMGDVEGLMEKVGSLG 308
Query 121 SAEKQQEMIERISKGKFTLQDMYDQFQNVLKMGPISKVMSMIP 163
EK E+++++ +G+FTL D YDQFQ +L +GPISK++ MIP
Sbjct 309 IDEK--EVVKKLRQGRFTLGDFYDQFQKILSLGPISKLLEMIP 349
> YPR088c
Length=541
Score = 176 bits (445), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 79/163 (48%), Positives = 112/163 (68%), Gaps = 0/163 (0%)
Query 1 SGRHRQEAALFEEMQQVADAVNPDDIVFVVDSHIGQACYDQAAAFAASVSVGSVIITKLD 60
SGRH QE LF+EM ++++ + P+ + V+D+ IGQA Q+ AF S G++I+TK+D
Sbjct 200 SGRHHQEEELFQEMIEISNVIKPNQTIMVLDASIGQAAEQQSKAFKESSDFGAIILTKMD 259
Query 61 GYAKGGGALSAVAATGAPIIFLGSGEHFDDFDPFDANSFVSRLLGLGDMGGLVNTLKEVI 120
G+A+GGGA+SAVAAT PIIF+G+GEH D + F SF+S+LLG+GD+ L L+ V
Sbjct 260 GHARGGGAISAVAATNTPIIFIGTGEHIHDLEKFSPKSFISKLLGIGDIESLFEQLQTVS 319
Query 121 SAEKQQEMIERISKGKFTLQDMYDQFQNVLKMGPISKVMSMIP 163
+ E + +E I KGKFTL D Q Q ++KMGP+S + MIP
Sbjct 320 NKEDAKATMENIQKGKFTLLDFKKQMQTIMKMGPLSNIAQMIP 362
> SPCC188.06c
Length=522
Score = 160 bits (404), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 83/163 (50%), Positives = 120/163 (73%), Gaps = 2/163 (1%)
Query 1 SGRHRQEAALFEEMQQVADAVNPDDIVFVVDSHIGQACYDQAAAFAASVSVGSVIITKLD 60
SGRH+QE LF EM +++DA+ PD + ++D+ IGQA Q+ AF + G+VIITKLD
Sbjct 192 SGRHQQEQELFAEMVEISDAIRPDQTIMILDASIGQAAESQSKAFKETADFGAVIITKLD 251
Query 61 GYAKGGGALSAVAATGAPIIFLGSGEHFDDFDPFDANSFVSRLLGLGDMGGLVNTLKEVI 120
G+AKGGGALSAVAAT PI+F+G+GEH +D + F SF+S+LLGLGD+ GL+ ++ +
Sbjct 252 GHAKGGGALSAVAATKTPIVFIGTGEHINDLERFSPRSFISKLLGLGDLEGLMEHVQSLD 311
Query 121 SAEKQQEMIERISKGKFTLQDMYDQFQNVLKMGPISKVMSMIP 163
+K M++ + +GKFT++D DQ N++K+GP+SK+ SMIP
Sbjct 312 FDKKN--MVKNLEQGKFTVRDFRDQLGNIMKLGPLSKMASMIP 352
> At5g03940
Length=564
Score = 112 bits (281), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 54/163 (33%), Positives = 90/163 (55%), Gaps = 0/163 (0%)
Query 1 SGRHRQEAALFEEMQQVADAVNPDDIVFVVDSHIGQACYDQAAAFAASVSVGSVIITKLD 60
+GR + + + +E++ V +NP +++ VVD+ GQ F + + I+TKLD
Sbjct 267 AGRLQIDKGMMDELKDVKKFLNPTEVLLVVDAMTGQEAAALVTTFNVEIGITGAILTKLD 326
Query 61 GYAKGGGALSAVAATGAPIIFLGSGEHFDDFDPFDANSFVSRLLGLGDMGGLVNTLKEVI 120
G ++GG ALS +G PI +G GE +D +PF + R+LG+GD+ V EV+
Sbjct 327 GDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVLSFVEKATEVM 386
Query 121 SAEKQQEMIERISKGKFTLQDMYDQFQNVLKMGPISKVMSMIP 163
E +++ ++I KF D Q + V KMG +++V+ MIP
Sbjct 387 RQEDAEDLQKKIMSAKFDFNDFLKQTRAVAKMGSMTRVLGMIP 429
> At2g45770
Length=373
Score = 74.7 bits (182), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 40/107 (37%), Positives = 57/107 (53%), Gaps = 6/107 (5%)
Query 1 SGRHRQEAALFEEMQQVADAVN------PDDIVFVVDSHIGQACYDQAAAFAASVSVGSV 54
SGR +L EE+ AV P++I+ V+D + G QA F V + +
Sbjct 263 SGRLHTNYSLMEELIACKKAVGKIVSGAPNEILLVLDGNTGLNMLPQAREFNEVVGITGL 322
Query 55 IITKLDGYAKGGGALSAVAATGAPIIFLGSGEHFDDFDPFDANSFVS 101
I+TKLDG A+GG +S V G P+ F+G GE +D PFD +FV+
Sbjct 323 ILTKLDGSARGGCVVSVVEELGIPVKFIGVGEAVEDLQPFDPEAFVN 369
> 7292603
Length=631
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 31/118 (26%), Positives = 58/118 (49%), Gaps = 11/118 (9%)
Query 1 SGRHRQEAALFEEMQQVADAVNPDDIVFVVDSHIGQACYDQAAAFAASVS---------- 50
+GR + L + ++ NPD ++FV ++ +G DQ F S++
Sbjct 499 AGRMQDNEPLMRSLSKLIKVNNPDLVLFVGEALVGNEAVDQLVKFNQSLADYSSNENPHI 558
Query 51 VGSVIITKLDGYA-KGGGALSAVAATGAPIIFLGSGEHFDDFDPFDANSFVSRLLGLG 107
+ +++TK D K G A+S TG PI+F+G+G+ + D + N+ V+ L+ +
Sbjct 559 IDGIVLTKFDTIDDKVGAAISMTYITGQPIVFVGTGQTYADLKAINVNAVVNSLMKIA 616
> At4g30600
Length=634
Score = 51.6 bits (122), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 30/115 (26%), Positives = 56/115 (48%), Gaps = 11/115 (9%)
Query 1 SGRHRQEAALFEEMQQVADAVNPDDIVFVVDSHIGQACYDQAAAFAASVS---------- 50
+GR + L + ++ + PD ++FV ++ +G DQ + F +S
Sbjct 519 AGRMQDNEPLMRALSKLINLNQPDLVLFVGEALVGNDAVDQLSKFNQKLSDLSTSGNPRL 578
Query 51 VGSVIITKLDGYA-KGGGALSAVAATGAPIIFLGSGEHFDDFDPFDANSFVSRLL 104
+ +++TK D K G ALS V +G+P++F+G G+ + D + + V LL
Sbjct 579 IDGILLTKFDTIDDKVGAALSMVYISGSPVMFVGCGQSYTDLKKLNVKAIVKTLL 633
> Hs4507223
Length=638
Score = 49.3 bits (116), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 29/115 (25%), Positives = 56/115 (48%), Gaps = 11/115 (9%)
Query 1 SGRHRQEAALFEEMQQVADAVNPDDIVFVVDSHIGQACYDQAAAFAASVS---------- 50
+GR + A L + ++ PD ++FV ++ +G DQ F +++
Sbjct 522 AGRMQDNAPLMTALAKLITVNTPDLVLFVGEALVGNEAVDQLVKFNRALADHSMAQTPRL 581
Query 51 VGSVIITKLDGYA-KGGGALSAVAATGAPIIFLGSGEHFDDFDPFDANSFVSRLL 104
+ +++TK D K G A+S T PI+F+G+G+ + D +A + V+ L+
Sbjct 582 IDGIVLTKFDTIDDKVGAAISMTYITSKPIVFVGTGQTYCDLRSLNAKAVVAALM 636
> Hs20481445
Length=638
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 29/115 (25%), Positives = 56/115 (48%), Gaps = 11/115 (9%)
Query 1 SGRHRQEAALFEEMQQVADAVNPDDIVFVVDSHIGQACYDQAAAFAASVS---------- 50
+GR + A L + ++ PD ++FV ++ +G DQ F +++
Sbjct 522 AGRMQDNAPLMTALAKLITVNTPDLVLFVGEALVGNEAVDQLVKFNRALADHSMAQTPRL 581
Query 51 VGSVIITKLDGYA-KGGGALSAVAATGAPIIFLGSGEHFDDFDPFDANSFVSRLL 104
+ +++TK D K G A+S T PI+F+G+G+ + D +A + V+ L+
Sbjct 582 IDGIVLTKFDTIDDKVGAAISMTYITSKPIVFVGTGQTYCDLRSLNAKAVVAALM 636
> YDR292c
Length=621
Score = 47.8 bits (112), Expect = 9e-06, Method: Composition-based stats.
Identities = 31/109 (28%), Positives = 51/109 (46%), Gaps = 5/109 (4%)
Query 1 SGRHRQEAALFEEMQQVADAVNPDDIVFVVDSHIGQACYDQAA----AFAASVSVGSVII 56
+GR + L ++ AD PD I+ V ++ +G QA AF ++ II
Sbjct 512 AGRRHNDPTLMSPLKSFADQAKPDKIIMVGEALVGTDSVQQAKNFNDAFGKGRNLDFFII 571
Query 57 TKLDGYAKGGGAL-SAVAATGAPIIFLGSGEHFDDFDPFDANSFVSRLL 104
+K D + G + + V ATG PI+F+G G+ + D V+ L+
Sbjct 572 SKCDTVGEMLGTMVNMVYATGIPILFVGVGQTYTDLRTLSVKWAVNTLM 620
> CE28419
Length=114
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 28/108 (25%), Positives = 50/108 (46%), Gaps = 12/108 (11%)
Query 10 LFEEMQQVADAVNPDDIVFVVDSHIGQACYDQAAAFAASVS-----------VGSVIITK 58
L E+ ++ PD ++FV ++ +G DQ F +++ + +++TK
Sbjct 7 LMRELAKLIRVNEPDLVLFVGEALVGNEAVDQLVKFNEALANHASPGQKPRLIDGIVLTK 66
Query 59 LDGYA-KGGGALSAVAATGAPIIFLGSGEHFDDFDPFDANSFVSRLLG 105
D K G A+S TG PI+F+G G+ + D + + V LL
Sbjct 67 FDTIDDKVGAAVSMTYITGQPIVFVGCGQTYSDLRNLNVGAVVHSLLN 114
> ECU10g0170
Length=425
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 27/112 (24%), Positives = 52/112 (46%), Gaps = 8/112 (7%)
Query 1 SGRHRQEAALFEEMQQVADAVNPDDIVFVVDSHIGQACYDQAAAFAASV-------SVGS 53
+GR + L + ++ PD I++V ++ +G D F ++ + S
Sbjct 312 AGRMHNKKNLMASLTKLMKINRPDHIIYVGEALVGSDSLDHIREFNKAIRDADQSRKIDS 371
Query 54 VIITKLDGYA-KGGGALSAVAATGAPIIFLGSGEHFDDFDPFDANSFVSRLL 104
+++TK+D + G L+ ++ API+FLG G+ D + VS L+
Sbjct 372 ILLTKVDTVDDRIGQILNMAFSSHAPILFLGVGQSNSDLSKIGSEDIVSSLM 423
> SPBC3B9.03
Length=547
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 29/108 (26%), Positives = 49/108 (45%), Gaps = 4/108 (3%)
Query 1 SGRHRQEAALFEEMQQVADAVNPDDIVFVVDSHIGQACYDQAAAFAASV---SVGSVIIT 57
+GR + L +++ A D I V ++ +G QA F AS+ + II+
Sbjct 439 AGRRHNDQRLMGSLEKFTKATKLDKIFQVAEALVGTDSLAQAKHFQASLYHRPLDGFIIS 498
Query 58 KLDGYAKGGGAL-SAVAATGAPIIFLGSGEHFDDFDPFDANSFVSRLL 104
K+D + G + V A PIIF+G G+ + D + V +L+
Sbjct 499 KVDTVGQLVGVMVGMVYAVRVPIIFVGIGQTYSDLRTLSVDWVVDQLM 546
> CE00643
Length=333
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 36/73 (49%), Gaps = 2/73 (2%)
Query 88 FDDFDPFDANSFVSRLL--GLGDMGGLVNTLKEVISAEKQQEMIERISKGKFTLQDMYDQ 145
F +++ F + ++++ LGD G ++ L+ SAEKQ+E + + DM
Sbjct 235 FVNYEQFRTSDAFTKVMEQNLGDRGIQLHGLEMCESAEKQEERFRNAGFKEVKVMDMNQI 294
Query 146 FQNVLKMGPISKV 158
F N L +S++
Sbjct 295 FNNFLDQKEVSRI 307
> CE29412
Length=1534
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 25/46 (54%), Gaps = 0/46 (0%)
Query 1 SGRHRQEAALFEEMQQVADAVNPDDIVFVVDSHIGQACYDQAAAFA 46
SG +Q +L + Q A+ V DD + VDSH+G+ ++ + A
Sbjct 740 SGGQKQRVSLARAVYQNAEIVLLDDPLSAVDSHVGKHIFENVISTA 785
> CE25015
Length=1528
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 25/46 (54%), Gaps = 0/46 (0%)
Query 1 SGRHRQEAALFEEMQQVADAVNPDDIVFVVDSHIGQACYDQAAAFA 46
SG +Q +L + Q A+ V DD + VDSH+G+ ++ + A
Sbjct 740 SGGQKQRVSLARAVYQNAEIVLLDDPLSAVDSHVGKHIFENVISTA 785
> 7292327
Length=717
Score = 28.9 bits (63), Expect = 4.6, Method: Composition-based stats.
Identities = 12/31 (38%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 123 EKQQEMIERISKGKFTLQDMYDQFQNVLKMG 153
+K E+IE +++ K + YDQF LK+G
Sbjct 403 KKTMELIEELTEDKENYKKFYDQFSKNLKLG 433
> SPAC23H3.14
Length=469
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 18/62 (29%), Positives = 27/62 (43%), Gaps = 1/62 (1%)
Query 70 SAVAATGAPIIFLGSGEHFDDFDPFDANSFVSRLLGLGDMGGLVNTLKEVISAEKQQEMI 129
S +A TG P+I G G F + P + + G NTL +++ K E+I
Sbjct 240 SLLAYTGFPLIIFGEGSMFSPYTPLQLVHVLEAKSSTSWLAGTTNTLI-MLNQNKMAEVI 298
Query 130 ER 131
R
Sbjct 299 VR 300
> CE07611
Length=933
Score = 28.5 bits (62), Expect = 6.7, Method: Composition-based stats.
Identities = 23/76 (30%), Positives = 41/76 (53%), Gaps = 4/76 (5%)
Query 84 SGEHFDDFDPFDA-NSFVSRLLGLGDMGGLV-NTLKEVISAEKQQEMIE-RISKGKF-TL 139
SGE FD D F+A + +L LG+ GL+ + +V + ++IE R+++ F
Sbjct 310 SGEDFDGMDRFEAREKVIQKLSELGNYNGLMKHEGAQVNICSRTGDIIEPRLTEQWFLDC 369
Query 140 QDMYDQFQNVLKMGPI 155
+DM+ + + +K G I
Sbjct 370 KDMFARSADAIKSGKI 385
> At2g17680
Length=292
Score = 28.5 bits (62), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 12/31 (38%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 130 ERISKGKFTLQDMYDQFQNVLKMGPISKVMS 160
E + G L+D+YD +++LKMG +V+S
Sbjct 46 ESVHSGLSGLEDLYDCSEDLLKMGSTQRVLS 76
Lambda K H
0.319 0.135 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2281134618
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40