bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2947_orf2
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
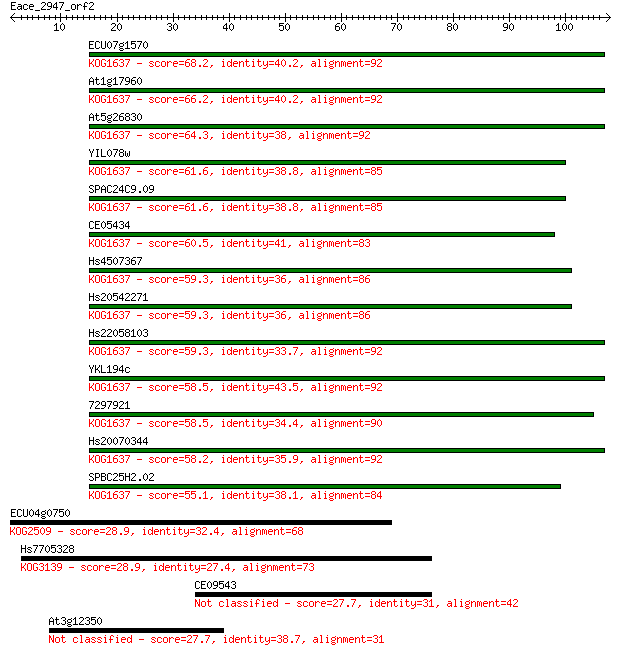
ECU07g1570 68.2 3e-12
At1g17960 66.2 1e-11
At5g26830 64.3 5e-11
YIL078w 61.6 4e-10
SPAC24C9.09 61.6 4e-10
CE05434 60.5 8e-10
Hs4507367 59.3 2e-09
Hs20542271 59.3 2e-09
Hs22058103 59.3 2e-09
YKL194c 58.5 3e-09
7297921 58.5 3e-09
Hs20070344 58.2 4e-09
SPBC25H2.02 55.1 3e-08
ECU04g0750 28.9 2.4
Hs7705328 28.9 2.7
CE09543 27.7 4.9
At3g12350 27.7 5.0
> ECU07g1570
Length=640
Score = 68.2 bits (165), Expect = 3e-12, Method: Composition-based stats.
Identities = 37/92 (40%), Positives = 51/92 (55%), Gaps = 3/92 (3%)
Query 15 EVPSALCGLLRMREFTQDDGHILCRMSQVLAEVTAFLLACQKLYKAVGFCKGEIVYHIST 74
E+ L GL R+R F QDD HI C QV E+ L +Y GF + E+V +ST
Sbjct 361 ELSGTLTGLTRVRRFQQDDAHIFCTKDQVKEEIKGCLEFLSFVYGVFGF-RFELV--LST 417
Query 75 RPKGSQGSEQEWARAEGLLRSALEQLDIPFTV 106
RP+ GS EW RAE L A+++ ++PF +
Sbjct 418 RPEKYLGSVDEWDRAEKALADAMDESNMPFKI 449
> At1g17960
Length=458
Score = 66.2 bits (160), Expect = 1e-11, Method: Composition-based stats.
Identities = 37/92 (40%), Positives = 49/92 (53%), Gaps = 3/92 (3%)
Query 15 EVPSALCGLLRMREFTQDDGHILCRMSQVLAEVTAFLLACQKLYKAVGFCKGEIVYHIST 74
E AL G+ R+R F QDD HI CR+ QV EV L +Y+ GF E+ +ST
Sbjct 184 EDSGALGGMTRVRRFVQDDAHIFCRVDQVEEEVKGVLDFIDYVYRIFGFTY-ELT--LST 240
Query 75 RPKGSQGSEQEWARAEGLLRSALEQLDIPFTV 106
RPK G + WA+AE L AL+ P+ +
Sbjct 241 RPKDHIGDLETWAKAENDLEKALDDFGKPWVI 272
> At5g26830
Length=676
Score = 64.3 bits (155), Expect = 5e-11, Method: Composition-based stats.
Identities = 35/92 (38%), Positives = 44/92 (47%), Gaps = 3/92 (3%)
Query 15 EVPSALCGLLRMREFTQDDGHILCRMSQVLAEVTAFLLACQKLYKAVGFCKGEIVYHIST 74
E AL GL R+R F QDD HI C QV EV L +YK GF +ST
Sbjct 405 EASGALSGLTRVRRFQQDDAHIFCTTEQVKGEVQGVLEFIDYVYKVFGF---TYELKLST 461
Query 75 RPKGSQGSEQEWARAEGLLRSALEQLDIPFTV 106
RP+ G + W +AE L+ A+E P +
Sbjct 462 RPEKYLGDLETWDKAEADLKEAIEAFGKPLVL 493
> YIL078w
Length=734
Score = 61.6 bits (148), Expect = 4e-10, Method: Composition-based stats.
Identities = 33/85 (38%), Positives = 43/85 (50%), Gaps = 3/85 (3%)
Query 15 EVPSALCGLLRMREFTQDDGHILCRMSQVLAEVTAFLLACQKLYKAVGFCKGEIVYHIST 74
E AL GL R+R F QDD HI C Q+ +E+ Q +Y GF E +ST
Sbjct 451 EFSGALSGLTRVRRFQQDDAHIFCTHDQIESEIENIFNFLQYIYGVFGF---EFKMELST 507
Query 75 RPKGSQGSEQEWARAEGLLRSALEQ 99
RP+ G + W AE L SAL++
Sbjct 508 RPEKYVGKIETWDAAESKLESALKK 532
> SPAC24C9.09
Length=473
Score = 61.6 bits (148), Expect = 4e-10, Method: Composition-based stats.
Identities = 33/85 (38%), Positives = 47/85 (55%), Gaps = 2/85 (2%)
Query 15 EVPSALCGLLRMREFTQDDGHILCRMSQVLAEVTAFLLACQKLYKAVGFCKGEIVYHIST 74
E AL GL R+R F QDDGHI C + E+ L +++Y +G K ++ ++ST
Sbjct 177 EASGALSGLTRLRCFHQDDGHIFCSPESIKDEIKNTLTFVKQVYSLLGMNKLKL--YLST 234
Query 75 RPKGSQGSEQEWARAEGLLRSALEQ 99
RP+ GS W AE LR AL++
Sbjct 235 RPEEHIGSLDTWNEAENGLREALQE 259
> CE05434
Length=725
Score = 60.5 bits (145), Expect = 8e-10, Method: Composition-based stats.
Identities = 34/84 (40%), Positives = 47/84 (55%), Gaps = 4/84 (4%)
Query 15 EVPSALCGLLRMREFTQDDGHILCRMSQVLAEVTAFLLACQKLYKAV-GFCKGEIVYHIS 73
E+ AL GL R+R F QDD HI CR Q+ E+ L + Y+ V GF ++S
Sbjct 443 EMSGALTGLTRVRRFQQDDAHIFCRQDQISEEIKQCLDFLEYAYEKVFGF---TFKLNLS 499
Query 74 TRPKGSQGSEQEWARAEGLLRSAL 97
TRP+G G+ + W +AE L +AL
Sbjct 500 TRPEGFLGNIETWDKAEADLTNAL 523
> Hs4507367
Length=712
Score = 59.3 bits (142), Expect = 2e-09, Method: Composition-based stats.
Identities = 31/86 (36%), Positives = 45/86 (52%), Gaps = 3/86 (3%)
Query 15 EVPSALCGLLRMREFTQDDGHILCRMSQVLAEVTAFLLACQKLYKAVGFCKGEIVYHIST 74
E+ AL GL R+R F QDD HI C M Q+ E+ L + +Y GF ++ST
Sbjct 432 ELSGALTGLTRVRRFQQDDAHIFCAMEQIEDEIKGCLDFLRTVYSVFGF---SFKLNLST 488
Query 75 RPKGSQGSEQEWARAEGLLRSALEQL 100
RP+ G + W +AE L ++L +
Sbjct 489 RPEKFLGDIEVWDQAEKQLENSLNEF 514
> Hs20542271
Length=723
Score = 59.3 bits (142), Expect = 2e-09, Method: Composition-based stats.
Identities = 31/86 (36%), Positives = 45/86 (52%), Gaps = 3/86 (3%)
Query 15 EVPSALCGLLRMREFTQDDGHILCRMSQVLAEVTAFLLACQKLYKAVGFCKGEIVYHIST 74
E+ AL GL R+R F QDD HI C M Q+ E+ L + +Y GF ++ST
Sbjct 444 ELSGALTGLTRVRRFQQDDAHIFCAMEQIEDEIKGCLDFLRTVYSVFGF---SFKLNLST 500
Query 75 RPKGSQGSEQEWARAEGLLRSALEQL 100
RP+ G + W +AE L ++L +
Sbjct 501 RPEKFLGDIEVWDQAEKQLENSLNEF 526
> Hs22058103
Length=707
Score = 59.3 bits (142), Expect = 2e-09, Method: Composition-based stats.
Identities = 31/92 (33%), Positives = 46/92 (50%), Gaps = 3/92 (3%)
Query 15 EVPSALCGLLRMREFTQDDGHILCRMSQVLAEVTAFLLACQKLYKAVGFCKGEIVYHIST 74
E+ L GL R+R F QDD HI C + Q+ E+ L Q +Y GF ++ST
Sbjct 428 ELSGTLSGLTRVRRFQQDDAHIFCTVEQIEEEIKGCLQFLQSVYSTFGF---SFQLNLST 484
Query 75 RPKGSQGSEQEWARAEGLLRSALEQLDIPFTV 106
RP+ G + W AE L+++L P+ +
Sbjct 485 RPENFLGEIEMWNEAEKQLQNSLMDFGEPWKM 516
> YKL194c
Length=462
Score = 58.5 bits (140), Expect = 3e-09, Method: Composition-based stats.
Identities = 40/99 (40%), Positives = 51/99 (51%), Gaps = 7/99 (7%)
Query 15 EVPSALCGLLRMREFTQDDGHILCRMSQVLAEVTAFLLACQKLY-KAVGFCKG----EIV 69
E AL GL R+R+F QDDGHI C SQV +E+ L +Y K F KG E
Sbjct 164 EASGALSGLTRLRKFHQDDGHIFCTPSQVKSEIFNSLKLIDIVYNKIFPFVKGGSGAESN 223
Query 70 YHI--STRPKGSQGSEQEWARAEGLLRSALEQLDIPFTV 106
Y I STRP G + W AE +L+ LE+ P+ +
Sbjct 224 YFINFSTRPDHFIGDLKVWNHAEQVLKEILEESGKPWKL 262
> 7297921
Length=690
Score = 58.5 bits (140), Expect = 3e-09, Method: Composition-based stats.
Identities = 31/90 (34%), Positives = 46/90 (51%), Gaps = 3/90 (3%)
Query 15 EVPSALCGLLRMREFTQDDGHILCRMSQVLAEVTAFLLACQKLYKAVGFCKGEIVYHIST 74
E+ AL GL R+R F QDD HI C Q+ +E+ L + +Y GF + +ST
Sbjct 413 ELSGALTGLTRVRRFQQDDAHIFCAPEQIKSEMKGCLEFLKYVYTIFGFSFQLV---LST 469
Query 75 RPKGSQGSEQEWARAEGLLRSALEQLDIPF 104
RP G ++W AE L +L + +P+
Sbjct 470 RPDNYLGELEQWNDAEKALAESLNEFGMPW 499
> Hs20070344
Length=718
Score = 58.2 bits (139), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 33/92 (35%), Positives = 48/92 (52%), Gaps = 3/92 (3%)
Query 15 EVPSALCGLLRMREFTQDDGHILCRMSQVLAEVTAFLLACQKLYKAVGFCKGEIVYHIST 74
E L GL R+R F QDD HI C Q+ AE+ + L + +Y +GF +ST
Sbjct 440 EASGGLGGLTRLRCFQQDDAHIFCTTDQLEAEIQSCLDFLRSVYAVLGFS---FRLALST 496
Query 75 RPKGSQGSEQEWARAEGLLRSALEQLDIPFTV 106
RP G G W +AE +L+ AL++ P+ +
Sbjct 497 RPSGFLGDPCLWDQAEQVLKQALKEFGEPWDL 528
> SPBC25H2.02
Length=703
Score = 55.1 bits (131), Expect = 3e-08, Method: Composition-based stats.
Identities = 32/87 (36%), Positives = 45/87 (51%), Gaps = 8/87 (9%)
Query 15 EVPSALCGLLRMREFTQDDGHILCRMSQVLAEVTAFLLACQKLYKAVGFCKGEIVYH--I 72
E AL GL R+R F QDD HI C QV +E+ +++Y GF +H +
Sbjct 412 EFSGALSGLTRVRRFQQDDAHIFCTPDQVRSEIEGCFDFLKEVYGTFGF-----TFHLEL 466
Query 73 STRPKGSQ-GSEQEWARAEGLLRSALE 98
STRP+ G W +AE L++AL+
Sbjct 467 STRPEEKYLGDLATWDKAEAQLKAALD 493
> ECU04g0750
Length=429
Score = 28.9 bits (63), Expect = 2.4, Method: Composition-based stats.
Identities = 22/93 (23%), Positives = 38/93 (40%), Gaps = 27/93 (29%)
Query 1 AALYIFHISLIVPREVPSALCG------------------LLRMREFTQDDGHILCRMSQ 42
AALY+ +VP+EVP CG L R+ +F + + ++C +
Sbjct 240 AALYM--DERMVPQEVPKKFCGQSLCFRKEAGAHGKDNAGLFRVHQFEKIEQFVICGPEE 297
Query 43 VLAEVTAFLLACQKLYKA-------VGFCKGEI 68
+ AC++ Y++ VG GE+
Sbjct 298 SQKYHEEMIKACEEFYQSLDISYNVVGIVSGEL 330
> Hs7705328
Length=227
Score = 28.9 bits (63), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 20/75 (26%), Positives = 39/75 (52%), Gaps = 7/75 (9%)
Query 3 LYIFHISLIVPREVPSALCGLLR-MREFTQDDGHILCRMSQVLAEVTAFLLACQKLYKAV 61
L +FH+S+ L+R + +F +D G+ S+V+ + + L+ LY+++
Sbjct 137 LQLFHLSVDNEHRGQGIAKALVRTVLQFARDQGY-----SEVVLDTSNIQLSAMGLYQSL 191
Query 62 GFCK-GEIVYHISTR 75
GF K G+ +H+ R
Sbjct 192 GFKKTGQSFFHVWAR 206
> CE09543
Length=347
Score = 27.7 bits (60), Expect = 4.9, Method: Composition-based stats.
Identities = 13/42 (30%), Positives = 21/42 (50%), Gaps = 0/42 (0%)
Query 34 GHILCRMSQVLAEVTAFLLACQKLYKAVGFCKGEIVYHISTR 75
G L + VL A LL+CQ Y+ V CK ++ ++ +
Sbjct 92 GEFLVCLHCVLTGFVASLLSCQFFYRYVALCKTHLLVYLQGK 133
> At3g12350
Length=455
Score = 27.7 bits (60), Expect = 5.0, Method: Composition-based stats.
Identities = 12/34 (35%), Positives = 18/34 (52%), Gaps = 3/34 (8%)
Query 8 ISLIVPREVPSALCGLLRMREFTQDDG---HILC 38
+SL+ P E+ S C R Q+DG H++C
Sbjct 20 LSLLTPAEISSFACTSKRFASLCQEDGKIWHVMC 53
Lambda K H
0.326 0.139 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164169380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40