bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2867_orf1
Length=122
Score E
Sequences producing significant alignments: (Bits) Value
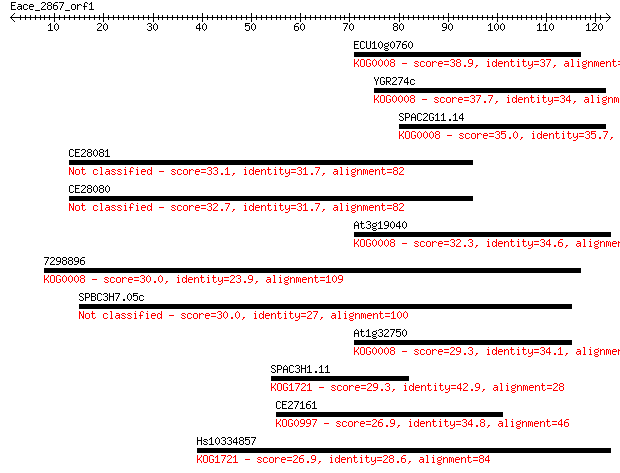
ECU10g0760 38.9 0.002
YGR274c 37.7 0.005
SPAC2G11.14 35.0 0.037
CE28081 33.1 0.14
CE28080 32.7 0.18
At3g19040 32.3 0.21
7298896 30.0 1.1
SPBC3H7.05c 30.0 1.1
At1g32750 29.3 1.8
SPAC3H1.11 29.3 2.0
CE27161 26.9 9.7
Hs10334857 26.9 9.8
> ECU10g0760
Length=883
Score = 38.9 bits (89), Expect = 0.002, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 71 PSVCFHDASDLTLSDDCPFVVFEHTEQQPFIINNAGMAIRIRRYVR 116
P A +LT+ D PF +FE++E++PF + N GM + Y R
Sbjct 302 PQGIIKKAGELTVRDASPFSLFEYSEEEPFFLVNPGMVSLLNIYYR 347
> YGR274c
Length=1066
Score = 37.7 bits (86), Expect = 0.005, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 75 FHDASDLTLSDDCPFVVFEHTEQQPFIINNAGMAIRIRRYVRPLQDQ 121
F + DLT+ D P + E++EQ P ++ GMA ++ Y R +Q
Sbjct 530 FSTSQDLTIGDTAPVYLMEYSEQTPVALSKFGMANKLINYYRKANEQ 576
> SPAC2G11.14
Length=979
Score = 35.0 bits (79), Expect = 0.037, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 26/42 (61%), Gaps = 0/42 (0%)
Query 80 DLTLSDDCPFVVFEHTEQQPFIINNAGMAIRIRRYVRPLQDQ 121
++T+ D ++ E +E+ P +++NAGMA RI Y R +Q
Sbjct 455 EITMGDTTHAILVEFSEEHPAVLSNAGMASRIVNYYRKKNEQ 496
> CE28081
Length=1311
Score = 33.1 bits (74), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 26/87 (29%), Positives = 37/87 (42%), Gaps = 12/87 (13%)
Query 13 LRMSFANCSKKRSTT---NNAERPWVVLPPFSSEVLRRSAESVAF--GGSSGFEDSSSSS 67
L+M FA + + N W V+ EV++ S E V + G S G E +
Sbjct 361 LKMVFAVVNDAGTVVLAHNEKLDSWSVIVNGCVEVVKPSGERVEYKLGDSFGAEPT---- 416
Query 68 AQHPSVCFHDASDLTLSDDCPFVVFEH 94
P+ H T+ DDC FV+ EH
Sbjct 417 ---PATQIHIGEMRTMVDDCEFVLVEH 440
> CE28080
Length=1470
Score = 32.7 bits (73), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 26/87 (29%), Positives = 37/87 (42%), Gaps = 12/87 (13%)
Query 13 LRMSFANCSKKRSTT---NNAERPWVVLPPFSSEVLRRSAESVAF--GGSSGFEDSSSSS 67
L+M FA + + N W V+ EV++ S E V + G S G E +
Sbjct 361 LKMVFAVVNDAGTVVLAHNEKLDSWSVIVNGCVEVVKPSGERVEYKLGDSFGAEPT---- 416
Query 68 AQHPSVCFHDASDLTLSDDCPFVVFEH 94
P+ H T+ DDC FV+ EH
Sbjct 417 ---PATQIHIGEMRTMVDDCEFVLVEH 440
> At3g19040
Length=1700
Score = 32.3 bits (72), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 30/54 (55%), Gaps = 3/54 (5%)
Query 71 PSVCFHDASDLTLSDDCPFVVFEHTEQQPFIINNAGMAIRIRRYVRPL--QDQH 122
P F SDL+ D F++ E+ E++P +++NAGM + Y + +DQH
Sbjct 592 PPGAFKKKSDLSNQDGHVFLM-EYCEERPLMLSNAGMGANLCTYYQKSSPEDQH 644
> 7298896
Length=2065
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 26/109 (23%), Positives = 46/109 (42%), Gaps = 18/109 (16%)
Query 8 FHRPDLRMSFANCSKKRSTTNNAERPWVVLPPFSSEVLRRSAESVAFGGSSGFEDSSSSS 67
FHRP L+ +++ +S + + +L + + +R E +A GG F
Sbjct 660 FHRPPLK-KYSHGPMAQSIPHPV---FPLLKTIAKKAKQREVERIASGGGDVF------- 708
Query 68 AQHPSVCFHDASDLTLSDDCPFVVFEHTEQQPFIINNAGMAIRIRRYVR 116
+ DL+ D V+ E E+ P +IN GM +I+ Y +
Sbjct 709 ------FMRNPEDLS-GRDGDIVLAEFCEEHPPLINQVGMCSKIKNYYK 750
> SPBC3H7.05c
Length=357
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 27/101 (26%), Positives = 41/101 (40%), Gaps = 6/101 (5%)
Query 15 MSFANCSKKRSTTNNAERPW-VVLPPFSSEVLRRSAESVAFGGSSGFEDSSSSSAQHPSV 73
+ N SK S + R W V P+ + ++A GS + +S S S
Sbjct 230 LMMGNVSKTTSVNDFWSRDWHVCTKPYFRVI--AWDPTIALTGS---KFLASCSCFFLSA 284
Query 74 CFHDASDLTLSDDCPFVVFEHTEQQPFIINNAGMAIRIRRY 114
FHD + ++S C F QPF+IN +R+Y
Sbjct 285 LFHDFAYWSISGRCSPAFFFQLLIQPFLINLEKRIPLLRKY 325
> At1g32750
Length=1919
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 25/44 (56%), Gaps = 1/44 (2%)
Query 71 PSVCFHDASDLTLSDDCPFVVFEHTEQQPFIINNAGMAIRIRRY 114
P F SDL+ D F++ E+ E++P +++NAGM + Y
Sbjct 753 PPGAFKKKSDLSTKDGHVFLM-EYCEERPLMLSNAGMGANLCTY 795
> SPAC3H1.11
Length=582
Score = 29.3 bits (64), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 12/28 (42%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 54 FGGSSGFEDSSSSSAQHPSVCFHDASDL 81
GG+S + +S S++QHPS ++D S L
Sbjct 101 IGGTSQYPSASFSTSQHPSQVYNDGSTL 128
> CE27161
Length=479
Score = 26.9 bits (58), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 22/46 (47%), Gaps = 3/46 (6%)
Query 55 GGSSGFEDSSSSSAQHPSVCFHDASDLTLSDDCPFVVFEHTEQQPF 100
GGS + S +S + V D + + D V+FEH EQ PF
Sbjct 7 GGSEAADTGSEASYKVSQVEIEDVEENLKNAD---VIFEHLEQLPF 49
> Hs10334857
Length=743
Score = 26.9 bits (58), Expect = 9.8, Method: Composition-based stats.
Identities = 24/97 (24%), Positives = 35/97 (36%), Gaps = 13/97 (13%)
Query 39 PFSSEVLRR------------SAESVAFGGSSGFE-DSSSSSAQHPSVCFHDASDLTLSD 85
P+SS ++R A S +F SSG + + + P C + S
Sbjct 558 PYSSHLIRHYRIHTGEKPYKCKACSKSFSDSSGLSVHRRTHTGEKPYTCKECGKAFSYSS 617
Query 86 DCPFVVFEHTEQQPFIINNAGMAIRIRRYVRPLQDQH 122
D HT Q+P+ G A R Y+ Q H
Sbjct 618 DVIQHRRIHTGQRPYKCEECGKAFNYRSYLTTHQRSH 654
Lambda K H
0.322 0.132 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194805952
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40