bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2866_orf1
Length=95
Score E
Sequences producing significant alignments: (Bits) Value
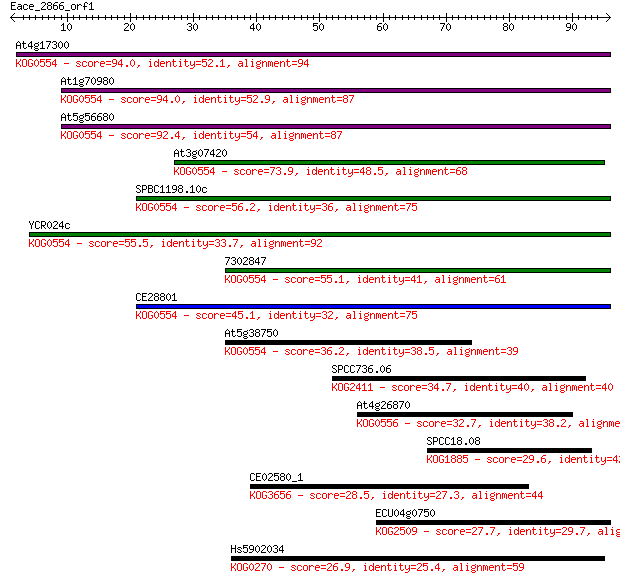
At4g17300 94.0 6e-20
At1g70980 94.0 7e-20
At5g56680 92.4 2e-19
At3g07420 73.9 7e-14
SPBC1198.10c 56.2 1e-08
YCR024c 55.5 2e-08
7302847 55.1 4e-08
CE28801 45.1 3e-05
At5g38750 36.2 0.014
SPCC736.06 34.7 0.046
At4g26870 32.7 0.18
SPCC18.08 29.6 1.3
CE02580_1 28.5 2.8
ECU04g0750 27.7 5.2
Hs5902034 26.9 9.9
> At4g17300
Length=567
Score = 94.0 bits (232), Expect = 6e-20, Method: Composition-based stats.
Identities = 49/94 (52%), Positives = 64/94 (68%), Gaps = 7/94 (7%)
Query 2 VESPADGQKIELQVADPQRGDSVEVLGGADAGKYPLAKKEHSREYLREIAHLRPRTQLIG 61
V S QK+EL+V + + V+G D+ YP+ KK SRE+LR AHLRPRT G
Sbjct 173 VASQGTKQKVELKV------EKIIVVGECDS-SYPIQKKRVSREFLRTKAHLRPRTNTFG 225
Query 62 AVARVRSQLAMATHRFFQDRGFLYVHTPIITTAD 95
AVARVR+ LA ATH+FFQ+ GF++V +PIIT +D
Sbjct 226 AVARVRNTLAYATHKFFQESGFVWVASPIITASD 259
> At1g70980
Length=571
Score = 94.0 bits (232), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 46/87 (52%), Positives = 59/87 (67%), Gaps = 6/87 (6%)
Query 9 QKIELQVADPQRGDSVEVLGGADAGKYPLAKKEHSREYLREIAHLRPRTQLIGAVARVRS 68
Q IEL V ++V +G D YPL K + + E+LR++ HLR RT LI AVAR+R+
Sbjct 119 QSIELSV------ETVIAVGTVDPTTYPLPKTKLTPEFLRDVLHLRSRTNLISAVARIRN 172
Query 69 QLAMATHRFFQDRGFLYVHTPIITTAD 95
LA ATH FFQ+ FLY+HTPIITT+D
Sbjct 173 ALAFATHSFFQEHSFLYIHTPIITTSD 199
> At5g56680
Length=572
Score = 92.4 bits (228), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 47/87 (54%), Positives = 58/87 (66%), Gaps = 6/87 (6%)
Query 9 QKIELQVADPQRGDSVEVLGGADAGKYPLAKKEHSREYLREIAHLRPRTQLIGAVARVRS 68
Q+IEL V V +G DA KYPL K + + E LR++ HLR RT I AVAR+R+
Sbjct 122 QQIELNVV------KVIDVGTVDASKYPLPKTKLTLETLRDVLHLRSRTNSISAVARIRN 175
Query 69 QLAMATHRFFQDRGFLYVHTPIITTAD 95
LA ATH FFQ+ FLY+HTPIITT+D
Sbjct 176 ALAFATHSFFQEHSFLYIHTPIITTSD 202
> At3g07420
Length=638
Score = 73.9 bits (180), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 33/68 (48%), Positives = 43/68 (63%), Gaps = 0/68 (0%)
Query 27 LGGADAGKYPLAKKEHSREYLREIAHLRPRTQLIGAVARVRSQLAMATHRFFQDRGFLYV 86
+G D KYPL+KK+ LR+ +H RPRT +G+V RV S L +A+H F Q GF YV
Sbjct 204 VGTVDPEKYPLSKKQLPLHMLRDFSHFRPRTTTVGSVTRVHSALTLASHTFLQYHGFQYV 263
Query 87 HTPIITTA 94
P+ITT
Sbjct 264 QVPVITTT 271
> SPBC1198.10c
Length=441
Score = 56.2 bits (134), Expect = 1e-08, Method: Composition-based stats.
Identities = 27/75 (36%), Positives = 42/75 (56%), Gaps = 0/75 (0%)
Query 21 GDSVEVLGGADAGKYPLAKKEHSREYLREIAHLRPRTQLIGAVARVRSQLAMATHRFFQD 80
+ +++ G + YP+ KK + E LR+I HLR RT G + R+RS A +FF
Sbjct 93 AEKIKIYGQINDDNYPIQKKHLTTEMLRQIPHLRLRTAKQGEIFRLRSDSLKALRQFFSS 152
Query 81 RGFLYVHTPIITTAD 95
+ F + PIIT++D
Sbjct 153 KDFTETNPPIITSSD 167
> YCR024c
Length=492
Score = 55.5 bits (132), Expect = 2e-08, Method: Composition-based stats.
Identities = 31/92 (33%), Positives = 50/92 (54%), Gaps = 3/92 (3%)
Query 4 SPADGQKIELQVADPQRGDSVEVLGGADAGKYPLAKKEHSREYLREIAHLRPRTQLIGAV 63
+P Q ELQ+ +P + S++++G + YPL KK + YLR + L+ RT + A+
Sbjct 97 TPNRKQPFELQIKNPVK--SIKLVGPV-SENYPLQKKYQTLRYLRSLPTLKYRTAYLSAI 153
Query 64 ARVRSQLAMATHRFFQDRGFLYVHTPIITTAD 95
R+RS + +FQ F V PI+T+ D
Sbjct 154 LRLRSFVEFQFMLYFQKNHFTKVSPPILTSND 185
> 7302847
Length=460
Score = 55.1 bits (131), Expect = 4e-08, Method: Composition-based stats.
Identities = 25/62 (40%), Positives = 38/62 (61%), Gaps = 1/62 (1%)
Query 35 YPLA-KKEHSREYLREIAHLRPRTQLIGAVARVRSQLAMATHRFFQDRGFLYVHTPIITT 93
YP + K++H EY+RE HLR R + A RVR + A H + D+ F+ ++TP++TT
Sbjct 108 YPFSPKQKHPPEYVREHLHLRSRVDFVAAQMRVRHKAQKAIHDYMDDQDFVQINTPLLTT 167
Query 94 AD 95
D
Sbjct 168 ND 169
> CE28801
Length=448
Score = 45.1 bits (105), Expect = 3e-05, Method: Composition-based stats.
Identities = 24/75 (32%), Positives = 39/75 (52%), Gaps = 4/75 (5%)
Query 21 GDSVEVLGGADAGKYPLAKKEHSREYLREIAHLRPRTQLIGAVARVRSQLAMATHRFFQD 80
+ ++V+G + +Y + S + LR+ HLR R A+ R RS + TH FF
Sbjct 80 AEQLKVVGNDENPRY----DDLSPDNLRKKTHLRARNSKFAALLRARSAIFRETHEFFMS 135
Query 81 RGFLYVHTPIITTAD 95
R F+++ TP +T D
Sbjct 136 RDFIHIDTPKLTRND 150
> At5g38750
Length=135
Score = 36.2 bits (82), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 25/39 (64%), Gaps = 0/39 (0%)
Query 35 YPLAKKEHSREYLREIAHLRPRTQLIGAVARVRSQLAMA 73
Y + K + + EYLR+ + R RT +I AV R+R ++ +A
Sbjct 89 YSIPKSKLTLEYLRDHTYFRARTSMIAAVTRIRHKMCLA 127
> SPCC736.06
Length=611
Score = 34.7 bits (78), Expect = 0.046, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 52 HLRPRTQLIGAVARVRSQLAMATHRFFQDRGFLYVHTPII 91
H++ R + R RS+LA H FF DR F V TP++
Sbjct 147 HIQLRAPKYNSYLRQRSRLAYQVHSFFNDREFCEVETPLL 186
> At4g26870
Length=504
Score = 32.7 bits (73), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 56 RTQLIGAVARVRSQLAMATHRFFQDRGFLYVHTP 89
RT A+ R++ Q+ +A + Q +GFL +HTP
Sbjct 209 RTPANQAIFRIQCQVQIAFREYLQSKGFLEIHTP 242
> SPCC18.08
Length=531
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 11/26 (42%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 67 RSQLAMATHRFFQDRGFLYVHTPIIT 92
R ++ + +FF +RGFL V TPI++
Sbjct 207 RYRIIESIRKFFSERGFLEVETPILS 232
> CE02580_1
Length=371
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 12/44 (27%), Positives = 24/44 (54%), Gaps = 0/44 (0%)
Query 39 KKEHSREYLREIAHLRPRTQLIGAVARVRSQLAMATHRFFQDRG 82
+++ +YLR+ A L+P L+ + +QL + H F ++G
Sbjct 76 RRQQKGKYLRDNASLQPLVWLLIVDVLMSAQLVVPIHNFLTEKG 119
> ECU04g0750
Length=429
Score = 27.7 bits (60), Expect = 5.2, Method: Composition-based stats.
Identities = 11/37 (29%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 59 LIGAVARVRSQLAMATHRFFQDRGFLYVHTPIITTAD 95
L G +AR+ L F +++G+ Y+ TP++ D
Sbjct 171 LSGKMARLAQALTRYAIDFLENKGYTYIQTPVMLRRD 207
> Hs5902034
Length=501
Score = 26.9 bits (58), Expect = 9.9, Method: Composition-based stats.
Identities = 15/59 (25%), Positives = 26/59 (44%), Gaps = 0/59 (0%)
Query 36 PLAKKEHSREYLREIAHLRPRTQLIGAVARVRSQLAMATHRFFQDRGFLYVHTPIITTA 94
P + + +Y RE ++P LI + Q + H + Q+ YVH I+ +A
Sbjct 128 PYVTLKDTEQYEREDFLIKPSDNLIVCGRAEQDQCNLEVHVYNQEEDSFYVHHDILLSA 186
Lambda K H
0.321 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1161385214
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40