bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2855_orf2
Length=120
Score E
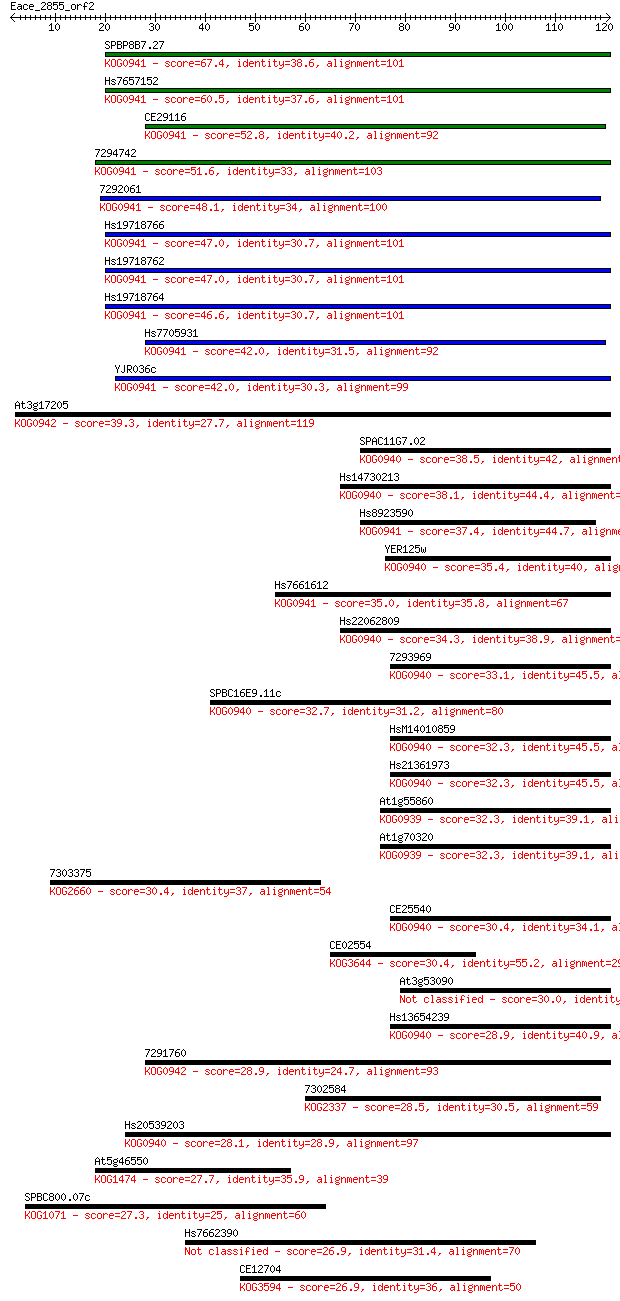
Sequences producing significant alignments: (Bits) Value
SPBP8B7.27 67.4 5e-12
Hs7657152 60.5 7e-10
CE29116 52.8 2e-07
7294742 51.6 4e-07
7292061 48.1 4e-06
Hs19718766 47.0 9e-06
Hs19718762 47.0 9e-06
Hs19718764 46.6 1e-05
Hs7705931 42.0 3e-04
YJR036c 42.0 3e-04
At3g17205 39.3 0.002
SPAC11G7.02 38.5 0.003
Hs14730213 38.1 0.004
Hs8923590 37.4 0.007
YER125w 35.4 0.024
Hs7661612 35.0 0.035
Hs22062809 34.3 0.063
7293969 33.1 0.12
SPBC16E9.11c 32.7 0.15
HsM14010859 32.3 0.20
Hs21361973 32.3 0.21
At1g55860 32.3 0.22
At1g70320 32.3 0.24
7303375 30.4 0.76
CE25540 30.4 0.78
CE02554 30.4 0.92
At3g53090 30.0 1.0
Hs13654239 28.9 2.2
7291760 28.9 2.6
7302584 28.5 2.9
Hs20539203 28.1 3.9
At5g46550 27.7 5.4
SPBC800.07c 27.3 7.2
Hs7662390 26.9 9.5
CE12704 26.9 9.7
> SPBP8B7.27
Length=807
Score = 67.4 bits (163), Expect = 5e-12, Method: Composition-based stats.
Identities = 39/101 (38%), Positives = 59/101 (58%), Gaps = 12/101 (11%)
Query 20 NTFSLVAHPFVLDASAKAEIIRMQATLEQQHQARQAEVQALLSLCRGGIGSPLPFLYLRV 79
N F +PF+L AK I+++ A + + +AR+A ++LS + P+L +RV
Sbjct 383 NCFCFCQYPFLLSMGAKISILQLDARRKMEIKAREAFFSSILSKM-----NVEPYLMIRV 437
Query 80 RRTHLVEDTLQQIASSSNSSSSTRTDLRKALKVKFIGEEGI 120
RR L+ED+L+QI + D RKALKV+F+GEEGI
Sbjct 438 RRDRLLEDSLRQI-------NDRNKDFRKALKVEFLGEEGI 471
> Hs7657152
Length=1050
Score = 60.5 bits (145), Expect = 7e-10, Method: Composition-based stats.
Identities = 38/105 (36%), Positives = 58/105 (55%), Gaps = 13/105 (12%)
Query 20 NTFSLVAHPFVLDASAKAEIIRMQATLEQQHQARQAEVQALLSLCRGGIGSPL----PFL 75
+T +L ++PF+ DA AK ++++ A L+ Q A +Q + L PL PFL
Sbjct 646 DTVTLCSYPFIFDAQAKTKMLQTDAELQMQVAVNGANLQNVFMLL---TLEPLLARSPFL 702
Query 76 YLRVRRTHLVEDTLQQIASSSNSSSSTRTDLRKALKVKFIGEEGI 120
L VRR +LV D L++++ S+ DL+K LKV F GEE +
Sbjct 703 VLHVRRNNLVGDALRELSIHSD------IDLKKPLKVIFDGEEAV 741
> CE29116
Length=1019
Score = 52.8 bits (125), Expect = 2e-07, Method: Composition-based stats.
Identities = 37/92 (40%), Positives = 51/92 (55%), Gaps = 10/92 (10%)
Query 28 PFVLDASAKAEIIRMQATLEQQHQARQAEVQALLSLCRGGIGSPLPFLYLRVRRTHLVED 87
PF+L+ AK E+I ++A L Q + RQ+ +L + G I LP L L VRR +V D
Sbjct 624 PFLLNGVAKGELIFVEAWLMQTLR-RQS---SLFNTAHGTIIFELPHLELTVRREFIVSD 679
Query 88 TLQQIASSSNSSSSTRTDLRKALKVKFIGEEG 119
T+ Q+A S S DL+K LKV GE+
Sbjct 680 TMNQMAGLSES------DLQKPLKVNIAGEDA 705
> 7294742
Length=973
Score = 51.6 bits (122), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 34/106 (32%), Positives = 58/106 (54%), Gaps = 8/106 (7%)
Query 18 HANTFSLVAHPFVLDASAKAEIIRMQATLEQQHQARQAEVQALLSLCRGGIG---SPLPF 74
HAN FS + + F+L S K + + + + + + R + + ++L+ G +G +P P
Sbjct 562 HANYFSFMLYAFILTPSTKVDALYYDSRM-RMYSERYSSLHSILNNI-GQVGQEDNPRPD 619
Query 75 LYLRVRRTHLVEDTLQQIASSSNSSSSTRTDLRKALKVKFIGEEGI 120
L L VRR L+ D L + + S+ DL+K L V+F+GE+GI
Sbjct 620 LKLTVRRDQLINDALIGLEMVAMSNPK---DLKKQLVVEFVGEQGI 662
> 7292061
Length=1057
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 34/101 (33%), Positives = 52/101 (51%), Gaps = 10/101 (9%)
Query 19 ANTFSLVAHPFVLDASAKAEIIRMQATLEQQHQARQAEVQALLSLCRGGIGSPLP-FLYL 77
A F++ + F+ D SAK +++ L Q H A S G+ P+ F+ L
Sbjct 656 ARDFNICNYSFIFDQSAKTALLQADQAL-QMHSAMANAATMAFSFFNYGM--PISQFIVL 712
Query 78 RVRRTHLVEDTLQQIASSSNSSSSTRTDLRKALKVKFIGEE 118
V R +LV+D+L+++ S S DL+K LK+KF GEE
Sbjct 713 NVTRENLVQDSLRELQHYSQS------DLKKPLKIKFHGEE 747
> Hs19718766
Length=875
Score = 47.0 bits (110), Expect = 9e-06, Method: Composition-based stats.
Identities = 31/101 (30%), Positives = 54/101 (53%), Gaps = 8/101 (7%)
Query 20 NTFSLVAHPFVLDASAKAEIIRMQATLEQQHQARQAEVQALLSLCRGGIGSPLPFLYLRV 79
N FS + PF+L+A K + + + R + L SL +G +P +L L+V
Sbjct 473 NKFSFMTCPFILNAVTKNLGLYYDNRIRMYSERR---ITVLYSLVQGQQLNP--YLRLKV 527
Query 80 RRTHLVEDTLQQIASSSNSSSSTRTDLRKALKVKFIGEEGI 120
RR H+++D L ++ + + + DL+K L V+F GE+G+
Sbjct 528 RRDHIIDDALVRLEMIAMENPA---DLKKQLYVEFEGEQGV 565
> Hs19718762
Length=852
Score = 47.0 bits (110), Expect = 9e-06, Method: Composition-based stats.
Identities = 31/101 (30%), Positives = 54/101 (53%), Gaps = 8/101 (7%)
Query 20 NTFSLVAHPFVLDASAKAEIIRMQATLEQQHQARQAEVQALLSLCRGGIGSPLPFLYLRV 79
N FS + PF+L+A K + + + R + L SL +G +P +L L+V
Sbjct 450 NKFSFMTCPFILNAVTKNLGLYYDNRIRMYSERR---ITVLYSLVQGQQLNP--YLRLKV 504
Query 80 RRTHLVEDTLQQIASSSNSSSSTRTDLRKALKVKFIGEEGI 120
RR H+++D L ++ + + + DL+K L V+F GE+G+
Sbjct 505 RRDHIIDDALVRLEMIAMENPA---DLKKQLYVEFEGEQGV 542
> Hs19718764
Length=872
Score = 46.6 bits (109), Expect = 1e-05, Method: Composition-based stats.
Identities = 31/101 (30%), Positives = 54/101 (53%), Gaps = 8/101 (7%)
Query 20 NTFSLVAHPFVLDASAKAEIIRMQATLEQQHQARQAEVQALLSLCRGGIGSPLPFLYLRV 79
N FS + PF+L+A K + + + R + L SL +G +P +L L+V
Sbjct 470 NKFSFMTCPFILNAVTKNLGLYYDNRIRMYSERR---ITVLYSLVQGQQLNP--YLRLKV 524
Query 80 RRTHLVEDTLQQIASSSNSSSSTRTDLRKALKVKFIGEEGI 120
RR H+++D L ++ + + + DL+K L V+F GE+G+
Sbjct 525 RRDHIIDDALVRLEMIAMENPA---DLKKQLYVEFEGEQGV 562
> Hs7705931
Length=1024
Score = 42.0 bits (97), Expect = 3e-04, Method: Composition-based stats.
Identities = 29/93 (31%), Positives = 46/93 (49%), Gaps = 7/93 (7%)
Query 28 PFVLDASAKAEIIRMQATLEQQHQARQAEVQ-ALLSLCRGGIGSPLPFLYLRVRRTHLVE 86
PF+ + +K +++ L+ + + +A ++ A + R + P L VRR HL+E
Sbjct 633 PFIFNNLSKIKLLHTDTLLKIESKKHKAYLRSAAIEEERESEFALRPTFDLTVRRNHLIE 692
Query 87 DTLQQIASSSNSSSSTRTDLRKALKVKFIGEEG 119
D L Q++ N DLRK L V F GE G
Sbjct 693 DVLNQLSQFENE------DLRKELWVSFSGEIG 719
> YJR036c
Length=892
Score = 42.0 bits (97), Expect = 3e-04, Method: Composition-based stats.
Identities = 30/99 (30%), Positives = 49/99 (49%), Gaps = 12/99 (12%)
Query 22 FSLVAHPFVLDASAKAEIIRMQATLEQQHQARQAEVQALLSLCRGGIGSPLPFLYLRVRR 81
FS +PF+L K I+ + +H+A QA L+SL +G S + ++VRR
Sbjct 473 FSFCKYPFILSLGIKISIMEYEIRRIMEHEAEQA---FLISLDKG--KSVDVYFKIKVRR 527
Query 82 THLVEDTLQQIASSSNSSSSTRTDLRKALKVKFIGEEGI 120
+ D+L+ I DL K+L+++F+ E GI
Sbjct 528 DVISHDSLRCIKEHQG-------DLLKSLRIEFVNEPGI 559
> At3g17205
Length=873
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 33/127 (25%), Positives = 59/127 (46%), Gaps = 30/127 (23%)
Query 2 TASSPF-----NHFALPQANLHAN--TFSLVAHPFVLDASAKAEIIRMQ-ATLEQQHQAR 53
T+SS F N + + QA + + L+ PF++ +++ +I Q AT Q H +
Sbjct 446 TSSSDFQADSVNEYFISQAIMEGTRANYILMQAPFLIPFTSRVKIFTTQLATARQSHGS- 504
Query 54 QAEVQALLSLCRGGIGSPLPFLYLRVRRTHLVEDTLQQIASSSNSSSSTRTDLRKALKVK 113
Q + + R R+RR H++ED Q+++ S DLR +++V
Sbjct 505 ----QGIFARNR-----------FRIRRDHILEDAYNQMSALSED------DLRSSIRVT 543
Query 114 FIGEEGI 120
F+ E G+
Sbjct 544 FVNELGV 550
> SPAC11G7.02
Length=767
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 21/51 (41%), Positives = 31/51 (60%), Gaps = 7/51 (13%)
Query 71 PLPF-LYLRVRRTHLVEDTLQQIASSSNSSSSTRTDLRKALKVKFIGEEGI 120
PLP +++VRR H+ ED+ +I S TDL+K L +KF GE+G+
Sbjct 408 PLPGQCHIKVRRNHIFEDSYAEIMRQS------ATDLKKRLMIKFDGEDGL 452
> Hs14730213
Length=1572
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 24/55 (43%), Positives = 28/55 (50%), Gaps = 7/55 (12%)
Query 67 GIGSPLPFLYLRVRRTHLVEDTLQQIASSSNSSSSTRTDL-RKALKVKFIGEEGI 120
G G L L +RR HL+ED QI S R DL R L V F+GEEG+
Sbjct 1208 GYGQGPGKLKLIIRRDHLLEDAFNQIMGYS------RKDLQRNKLYVTFVGEEGL 1256
> Hs8923590
Length=379
Score = 37.4 bits (85), Expect = 0.007, Method: Composition-based stats.
Identities = 21/47 (44%), Positives = 27/47 (57%), Gaps = 6/47 (12%)
Query 71 PLPFLYLRVRRTHLVEDTLQQIASSSNSSSSTRTDLRKALKVKFIGE 117
P P LRVRR+ LV+D L+Q+ S + TD K L V+FI E
Sbjct 25 PSPRFILRVRRSRLVKDALRQL------SQAEATDFCKVLVVEFINE 65
> YER125w
Length=809
Score = 35.4 bits (80), Expect = 0.024, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 27/45 (60%), Gaps = 6/45 (13%)
Query 76 YLRVRRTHLVEDTLQQIASSSNSSSSTRTDLRKALKVKFIGEEGI 120
+++VRR ++ ED Q+I T DL+K L +KF GEEG+
Sbjct 456 HIKVRRKNIFEDAYQEIMRQ------TPEDLKKRLMIKFDGEEGL 494
> Hs7661612
Length=110
Score = 35.0 bits (79), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 35/67 (52%), Gaps = 6/67 (8%)
Query 54 QAEVQALLSLCRGGIGSPLPFLYLRVRRTHLVEDTLQQIASSSNSSSSTRTDLRKALKVK 113
QA Q + SL I S P L L VRR ++V D ++ + + N D +K LKV
Sbjct 5 QAHRQNVSSLFLPVIESVNPCLILVVRRENIVGDAMEVLRKAKN------IDYKKPLKVI 58
Query 114 FIGEEGI 120
F+GE+ +
Sbjct 59 FVGEDAV 65
> Hs22062809
Length=1606
Score = 34.3 bits (77), Expect = 0.063, Method: Compositional matrix adjust.
Identities = 21/55 (38%), Positives = 29/55 (52%), Gaps = 7/55 (12%)
Query 67 GIGSPLPFLYLRVRRTHLVEDTLQQIASSSNSSSSTRTDL-RKALKVKFIGEEGI 120
G G + L +RR HL+E T Q+ + S R +L R L V F+GEEG+
Sbjct 1242 GFGQGPGKIKLIIRRDHLLEGTFNQVMAYS------RKELQRNKLYVTFVGEEGL 1290
> 7293969
Length=1082
Score = 33.1 bits (74), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 20/45 (44%), Positives = 29/45 (64%), Gaps = 7/45 (15%)
Query 77 LRVRRTHLVEDTLQQIASSSNSSSSTRTDLRKA-LKVKFIGEEGI 120
+R+RRT ++ED+ + I SS T+TDL K L V+F GE G+
Sbjct 727 IRIRRTSILEDSYRII------SSVTKTDLLKTKLWVEFEGETGL 765
> SPBC16E9.11c
Length=786
Score = 32.7 bits (73), Expect = 0.15, Method: Composition-based stats.
Identities = 25/82 (30%), Positives = 41/82 (50%), Gaps = 8/82 (9%)
Query 41 RMQATLEQQHQARQAEVQALLSLCRGGIG-SPLPF-LYLRVRRTHLVEDTLQQIASSSNS 98
R+ + L+Q + + + L R G PLP ++VRR H+ ED+ +I S
Sbjct 396 RLPSALDQDVPQYKCDFRRKLIYFRSQPGMRPLPGQCNVKVRRDHIFEDSYAEIMRYSAH 455
Query 99 SSSTRTDLRKALKVKFIGEEGI 120
DL+K L ++F GE+G+
Sbjct 456 ------DLKKRLMIRFDGEDGL 471
> HsM14010859
Length=862
Score = 32.3 bits (72), Expect = 0.20, Method: Composition-based stats.
Identities = 20/44 (45%), Positives = 24/44 (54%), Gaps = 6/44 (13%)
Query 77 LRVRRTHLVEDTLQQIASSSNSSSSTRTDLRKALKVKFIGEEGI 120
+ V R L ED+ QQI S S DLR+ L V F GEEG+
Sbjct 509 ITVTRKTLFEDSFQQIMSFSPQ------DLRRRLWVIFPGEEGL 546
> Hs21361973
Length=903
Score = 32.3 bits (72), Expect = 0.21, Method: Composition-based stats.
Identities = 20/44 (45%), Positives = 24/44 (54%), Gaps = 6/44 (13%)
Query 77 LRVRRTHLVEDTLQQIASSSNSSSSTRTDLRKALKVKFIGEEGI 120
+ V R L ED+ QQI S S DLR+ L V F GEEG+
Sbjct 550 ITVTRKTLFEDSFQQIMSFSPQ------DLRRRLWVIFPGEEGL 587
> At1g55860
Length=3891
Score = 32.3 bits (72), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 26/46 (56%), Gaps = 6/46 (13%)
Query 75 LYLRVRRTHLVEDTLQQIASSSNSSSSTRTDLRKALKVKFIGEEGI 120
L + VRR +++ED+ Q+ S DL+ L V+F GEEGI
Sbjct 3529 LRISVRRAYVLEDSYNQLRMRSPQ------DLKGRLNVQFQGEEGI 3568
> At1g70320
Length=3658
Score = 32.3 bits (72), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 26/46 (56%), Gaps = 6/46 (13%)
Query 75 LYLRVRRTHLVEDTLQQIASSSNSSSSTRTDLRKALKVKFIGEEGI 120
L + VRR +++ED+ Q+ S DL+ L V+F GEEGI
Sbjct 3296 LRISVRRAYVLEDSYNQLRMRSPQ------DLKGRLNVQFQGEEGI 3335
> 7303375
Length=1368
Score = 30.4 bits (67), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 34/60 (56%), Gaps = 7/60 (11%)
Query 9 HFALPQANLHANTFSLVAH---PFVLDASAKAEIIRMQATLEQQHQARQA---EVQALLS 62
HFA+PQA H N + + + P + ASA ++ + +L++ H A+QA + QA+ S
Sbjct 713 HFAVPQAPTHRNMYHMQRYQSTPSSI-ASAANKMPKRSMSLDESHPAKQARLSQAQAMAS 771
> CE25540
Length=817
Score = 30.4 bits (67), Expect = 0.78, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 25/44 (56%), Gaps = 6/44 (13%)
Query 77 LRVRRTHLVEDTLQQIASSSNSSSSTRTDLRKALKVKFIGEEGI 120
+ V R ++ ED+ Q+I + DLR+ L ++F GEEG+
Sbjct 441 ITVSRNNVFEDSFQEIMRKN------AVDLRRRLYIQFRGEEGL 478
> CE02554
Length=505
Score = 30.4 bits (67), Expect = 0.92, Method: Composition-based stats.
Identities = 16/32 (50%), Positives = 21/32 (65%), Gaps = 3/32 (9%)
Query 65 RGGIGSPLPFLYLRVR---RTHLVEDTLQQIA 93
RG +G+ L+LR+R R HLVED +QIA
Sbjct 317 RGDLGAHARHLFLRLRTNERQHLVEDPPEQIA 348
> At3g53090
Length=1142
Score = 30.0 bits (66), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 24/42 (57%), Gaps = 7/42 (16%)
Query 79 VRRTHLVEDTLQQIASSSNSSSSTRTDLRKALKVKFIGEEGI 120
VRR H+VED QQ+ +S + L+ ++ V F+ E G+
Sbjct 778 VRRGHVVEDGFQQL-------NSIGSRLKSSIHVSFVNESGL 812
> Hs13654239
Length=922
Score = 28.9 bits (63), Expect = 2.2, Method: Composition-based stats.
Identities = 18/44 (40%), Positives = 23/44 (52%), Gaps = 6/44 (13%)
Query 77 LRVRRTHLVEDTLQQIASSSNSSSSTRTDLRKALKVKFIGEEGI 120
+ V R L ED+ QQI + DLR+ L V F GEEG+
Sbjct 569 INVSRQTLFEDSFQQIMALKP------YDLRRRLYVIFRGEEGL 606
> 7291760
Length=1122
Score = 28.9 bits (63), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 23/93 (24%), Positives = 42/93 (45%), Gaps = 17/93 (18%)
Query 28 PFVLDASAKAEIIRMQATLEQQHQARQAEVQALLSLCRGGIGSPLPFLYLRVRRTHLVED 87
PFV+ + + I +Q+ + Q +QA L +G P + + VRR+HL ED
Sbjct 728 PFVVPFNKRVSI--LQSLVAASKMRVQGNMQAFL---QG------PSVLITVRRSHLYED 776
Query 88 TLQQIASSSNSSSSTRTDLRKALKVKFIGEEGI 120
++ + DLR +++F+ G+
Sbjct 777 AYDKLRPDNEP------DLRFKFRIQFVSSLGL 803
> 7302584
Length=684
Score = 28.5 bits (62), Expect = 2.9, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 29/59 (49%), Gaps = 5/59 (8%)
Query 60 LLSLCRGGIGSPLPFLYLRVRRTHLVEDTLQQIASSSNSSSSTRTDLRKALKVKFIGEE 118
LL C +G PL FL LR + ++D+L + + DL ++ +KF+G E
Sbjct 236 LLQQCPSFVGKPLKFLGLRHNQQMNIDDSLVWKVIQTEA-----CDLSQSENIKFVGWE 289
> Hs20539203
Length=757
Score = 28.1 bits (61), Expect = 3.9, Method: Composition-based stats.
Identities = 28/97 (28%), Positives = 41/97 (42%), Gaps = 27/97 (27%)
Query 24 LVAHPFVLDASAKAEIIRMQATLEQQHQARQAEVQALLSLCRGGIGSPLPFLYLRVRRTH 83
L A + D K +++R + +L QQ QA CR + V R
Sbjct 369 LPAQRYERDLVQKLKVLRHELSL-QQPQAGH---------CR-----------IEVSREE 407
Query 84 LVEDTLQQIASSSNSSSSTRTDLRKALKVKFIGEEGI 120
+ E++ +QI DL+K L VKF GEEG+
Sbjct 408 IFEESYRQIMKMRPK------DLKKRLMVKFRGEEGL 438
> At5g46550
Length=506
Score = 27.7 bits (60), Expect = 5.4, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 18 HANTFSLVAHPFVLDASAKAEIIRMQATLEQQHQARQAE 56
+A T H VL + KA++IR+Q EQ +A++ E
Sbjct 360 YAGTIIKAKHRIVLGQNNKADLIRIQIEKEQMERAQREE 398
> SPBC800.07c
Length=299
Score = 27.3 bits (59), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 15/60 (25%), Positives = 32/60 (53%), Gaps = 0/60 (0%)
Query 4 SSPFNHFALPQANLHANTFSLVAHPFVLDASAKAEIIRMQATLEQQHQARQAEVQALLSL 63
++PF A A+ + + ++A+ K EI++ QA + + H+A + +V + +SL
Sbjct 121 TTPFQDLARRIASTFLHYLPTNHSSYSVEATLKNEILKHQAYVSKNHEANEKDVSSNVSL 180
> Hs7662390
Length=988
Score = 26.9 bits (58), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 37/73 (50%), Gaps = 9/73 (12%)
Query 36 KAEIIRMQATLEQ---QHQARQAEVQALLSLCRGGIGSPLPFLYLRVRRTHLVEDTLQQI 92
++E+I ++ T+E Q+ A QA +Q L+ G P LR+RR H E
Sbjct 249 ESELIELRETIEMLKAQNSAAQAAIQGALN------GPDHPPKDLRIRRQHSSESVSSIN 302
Query 93 ASSSNSSSSTRTD 105
+++S+SS + D
Sbjct 303 SATSHSSIGSGND 315
> CE12704
Length=2163
Score = 26.9 bits (58), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 18/55 (32%), Positives = 26/55 (47%), Gaps = 5/55 (9%)
Query 47 EQQHQARQAEVQALLSLCRGGIGSPLPFLYLRVRRTHLVEDT-----LQQIASSS 96
+ H AR A + +SL PF L V H++EDT LQ+I +S+
Sbjct 1728 DHGHPARAAVGKVQVSLKMSSWSGSAPFFVLPVYEVHVLEDTSVGTVLQKIRASN 1782
Lambda K H
0.319 0.129 0.354
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160968786
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40