bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2778_orf1
Length=77
Score E
Sequences producing significant alignments: (Bits) Value
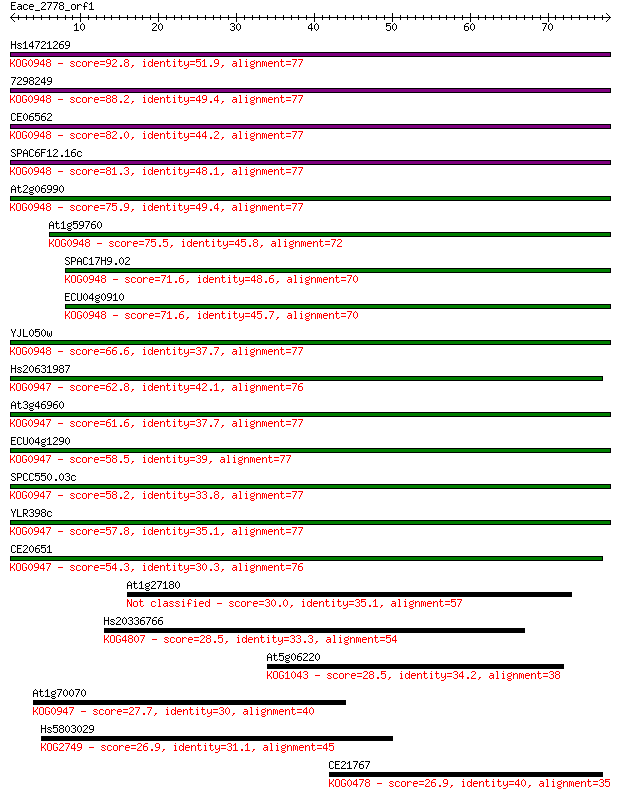
Hs14721269 92.8 1e-19
7298249 88.2 3e-18
CE06562 82.0 2e-16
SPAC6F12.16c 81.3 4e-16
At2g06990 75.9 2e-14
At1g59760 75.5 2e-14
SPAC17H9.02 71.6 3e-13
ECU04g0910 71.6 4e-13
YJL050w 66.6 1e-11
Hs20631987 62.8 2e-10
At3g46960 61.6 3e-10
ECU04g1290 58.5 3e-09
SPCC550.03c 58.2 4e-09
YLR398c 57.8 5e-09
CE20651 54.3 6e-08
At1g27180 30.0 1.2
Hs20336766 28.5 3.4
At5g06220 28.5 3.6
At1g70070 27.7 5.4
Hs5803029 26.9 9.2
CE21767 26.9 9.9
> Hs14721269
Length=1042
Score = 92.8 bits (229), Expect = 1e-19, Method: Composition-based stats.
Identities = 40/77 (51%), Positives = 57/77 (74%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
L+ + Y WA G +FA + + T ++EG++IRCMRRLEELLRQ+ AAKAIGN ELE K
Sbjct 966 LMDVVYTWATGATFAHICKMTDVFEGSIIRCMRRLEELLRQMCQAAKAIGNTELENKFAE 1025
Query 61 AVRQLRRGSIFSSSLYL 77
+ +++R +F++SLYL
Sbjct 1026 GITKIKRDIVFAASLYL 1042
> 7298249
Length=1055
Score = 88.2 bits (217), Expect = 3e-18, Method: Composition-based stats.
Identities = 38/77 (49%), Positives = 55/77 (71%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
L+ + W +G SF A+ + T ++EG++IRCMRRLEELLRQ+ A+K IGN +LE K
Sbjct 979 LMDVVLAWCKGSSFLAVCKMTDIFEGSIIRCMRRLEELLRQMCQASKTIGNTDLENKFSE 1038
Query 61 AVRQLRRGSIFSSSLYL 77
+R L+R +F++SLYL
Sbjct 1039 GIRLLKRDIVFAASLYL 1055
> CE06562
Length=1026
Score = 82.0 bits (201), Expect = 2e-16, Method: Composition-based stats.
Identities = 34/77 (44%), Positives = 58/77 (75%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
L+ + Y W G +F+ ++++T ++EG++IR +RRLEE+LR++ +AAKA+ N+ELE K
Sbjct 950 LMDVVYQWVNGATFSEIVKTTDVFEGSIIRTLRRLEEVLREMINAAKALANKELEQKFED 1009
Query 61 AVRQLRRGSIFSSSLYL 77
A + L+R +F++SLYL
Sbjct 1010 ARKNLKRDIVFAASLYL 1026
> SPAC6F12.16c
Length=1117
Score = 81.3 bits (199), Expect = 4e-16, Method: Composition-based stats.
Identities = 37/77 (48%), Positives = 51/77 (66%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
L+ + Y WA G SFA + + T +YEG++IR RRLEEL+RQ+ AAK IGN L+ K
Sbjct 1041 LMEVVYAWAHGASFAQICKMTDVYEGSLIRMFRRLEELIRQMVDAAKVIGNTSLQQKMED 1100
Query 61 AVRQLRRGSIFSSSLYL 77
+ + R +FS+SLYL
Sbjct 1101 TIACIHRDIVFSASLYL 1117
> At2g06990
Length=996
Score = 75.9 bits (185), Expect = 2e-14, Method: Composition-based stats.
Identities = 38/89 (42%), Positives = 56/89 (62%), Gaps = 12/89 (13%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLR------------QLASAAKA 48
L+ + Y W++G SFA +I+ T ++EG++IR RRL+E L QL +AA+A
Sbjct 908 LMDVIYSWSKGASFAEIIQMTDIFEGSIIRSARRLDEFLNQKSDDTLIGDFLQLRAAAEA 967
Query 49 IGNQELEAKCVAAVRQLRRGSIFSSSLYL 77
+G LE+K AA LRRG +F++SLYL
Sbjct 968 VGESSLESKFAAASESLRRGIMFANSLYL 996
> At1g59760
Length=988
Score = 75.5 bits (184), Expect = 2e-14, Method: Composition-based stats.
Identities = 33/72 (45%), Positives = 52/72 (72%), Gaps = 0/72 (0%)
Query 6 YGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVAAVRQL 65
Y WA+G F ++ ++EG++IR +RR+EE+L+QL AAK+IG +LEAK AV ++
Sbjct 917 YAWAKGSKFYEVMEIARVFEGSLIRAIRRMEEVLQQLIVAAKSIGETQLEAKLEEAVSKI 976
Query 66 RRGSIFSSSLYL 77
+R +F++SLYL
Sbjct 977 KRDIVFAASLYL 988
> SPAC17H9.02
Length=1030
Score = 71.6 bits (174), Expect = 3e-13, Method: Composition-based stats.
Identities = 34/70 (48%), Positives = 46/70 (65%), Gaps = 0/70 (0%)
Query 8 WAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVAAVRQLRR 67
W G SF + + LYEG+++R RRL+ELL+QL AA +GN EL+ K V ++L R
Sbjct 961 WINGASFQEICIVSKLYEGSIVRTFRRLDELLKQLEHAAIVLGNNELKEKSVLTEQKLHR 1020
Query 68 GSIFSSSLYL 77
IFS+SLYL
Sbjct 1021 DIIFSASLYL 1030
> ECU04g0910
Length=933
Score = 71.6 bits (174), Expect = 4e-13, Method: Composition-based stats.
Identities = 32/70 (45%), Positives = 50/70 (71%), Gaps = 0/70 (0%)
Query 8 WAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVAAVRQLRR 67
W G +F ++ TS++EG++IR +RLEELLRQL+SAA+ IGN ELE + +++R
Sbjct 864 WVCGCTFISICSKTSIFEGSIIRTFKRLEELLRQLSSAARVIGNTELENMFALGIVKIKR 923
Query 68 GSIFSSSLYL 77
+F++SLY+
Sbjct 924 DIVFANSLYI 933
> YJL050w
Length=1073
Score = 66.6 bits (161), Expect = 1e-11, Method: Composition-based stats.
Identities = 29/77 (37%), Positives = 48/77 (62%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
L+ + Y W +G +F + + T +YEG++IR +RLEEL+++L A IGN L+ K A
Sbjct 997 LMEVVYEWCRGATFTQICKMTDVYEGSLIRMFKRLEELVKELVDVANTIGNSSLKEKMEA 1056
Query 61 AVRQLRRGSIFSSSLYL 77
++ + R + + SLYL
Sbjct 1057 VLKLIHRDIVSAGSLYL 1073
> Hs20631987
Length=1246
Score = 62.8 bits (151), Expect = 2e-10, Method: Composition-based stats.
Identities = 32/76 (42%), Positives = 45/76 (59%), Gaps = 0/76 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
LV + Y WA+G F+ L + EG V+RC++RL E+ R L AA+ +G L AK
Sbjct 1169 LVEVVYEWARGMPFSELAGLSGTPEGLVVRCIQRLAEMCRSLRGAARLVGEPVLGAKMET 1228
Query 61 AVRQLRRGSIFSSSLY 76
A LRR +F++SLY
Sbjct 1229 AATLLRRDIVFAASLY 1244
> At3g46960
Length=1347
Score = 61.6 bits (148), Expect = 3e-10, Method: Composition-based stats.
Identities = 29/77 (37%), Positives = 46/77 (59%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
LV + Y WA+G FA + T + EG ++R + RL+E R+ +AA +GN L K A
Sbjct 1268 LVEVVYEWAKGTPFAEICELTDVPEGLIVRTIVRLDETCREFKNAAAIMGNSALHKKMDA 1327
Query 61 AVRQLRRGSIFSSSLYL 77
A ++R +F++SLY+
Sbjct 1328 ASNAIKRDIVFAASLYV 1344
> ECU04g1290
Length=881
Score = 58.5 bits (140), Expect = 3e-09, Method: Composition-based stats.
Identities = 30/77 (38%), Positives = 42/77 (54%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
LV Y W G S ++ ++ EGT +R + RLEE R+L S + IG++ LE K
Sbjct 805 LVDAVYDWCNGSSLGKIVSRYNVLEGTFVRLVLRLEECCRELISVSTMIGDKSLEEKIGD 864
Query 61 AVRQLRRGSIFSSSLYL 77
A ++R IF SLYL
Sbjct 865 ASASMKRDIIFLPSLYL 881
> SPCC550.03c
Length=1213
Score = 58.2 bits (139), Expect = 4e-09, Method: Composition-based stats.
Identities = 26/77 (33%), Positives = 45/77 (58%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
L+ + Y WA+G SF + T + EG+++R + RL+E+LR+ AA+ +G+ + K
Sbjct 1137 LMEVCYEWARGMSFNRITDLTDVLEGSIVRTIIRLDEVLRECRGAARVVGDSSMYTKMEE 1196
Query 61 AVRQLRRGSIFSSSLYL 77
+RR +F SLY+
Sbjct 1197 CQNLIRRNIVFCPSLYM 1213
> YLR398c
Length=1287
Score = 57.8 bits (138), Expect = 5e-09, Method: Composition-based stats.
Identities = 27/77 (35%), Positives = 47/77 (61%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
++ + Y WA+G SF ++ + EGTV+R + L+E+ R++ +A+ IGN L K
Sbjct 1211 MMNVVYEWARGLSFKEIMEMSPEAEGTVVRVITWLDEICREVKTASIIIGNSTLHMKMSR 1270
Query 61 AVRQLRRGSIFSSSLYL 77
A ++R +F++SLYL
Sbjct 1271 AQELIKRDIVFAASLYL 1287
> CE20651
Length=1260
Score = 54.3 bits (129), Expect = 6e-08, Method: Composition-based stats.
Identities = 23/76 (30%), Positives = 44/76 (57%), Gaps = 0/76 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
L+ + Y WA G F ++ T EG +++C++RL+E+ + + +A + +G+ L K
Sbjct 1182 LMEVVYEWANGTPFYQIMEMTDCQEGLIVKCIQRLDEVCKDVRNAGRIVGDPALVEKMEE 1241
Query 61 AVRQLRRGSIFSSSLY 76
+RR +F++SLY
Sbjct 1242 VSASIRRDIVFAASLY 1257
> At1g27180
Length=1556
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 31/58 (53%), Gaps = 7/58 (12%)
Query 16 ALIRSTSLY-EGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVAAVRQLRRGSIFS 72
AL+ TS Y EG++I R E+L +L+ NQ+ E KC+ + L+ S +S
Sbjct 484 ALVGETSWYGEGSLIVITTRDSEILSKLSV------NQQYEVKCLTEPQALKLFSFYS 535
> Hs20336766
Length=593
Score = 28.5 bits (62), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 24/54 (44%), Gaps = 9/54 (16%)
Query 13 SFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVAAVRQLR 66
AL+R E T+ RC + +ELLR NQEL + + QLR
Sbjct 443 EIGALMRQAEEREHTLRRCQQEGQELLRH---------NQELHGRLSEEIDQLR 487
> At5g06220
Length=813
Score = 28.5 bits (62), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 13/40 (32%), Positives = 28/40 (70%), Gaps = 2/40 (5%)
Query 34 RLEELLRQLASAAKAIGNQELEAKC--VAAVRQLRRGSIF 71
RLE LL+QL +++ + G ++++A C + +R+L++ + F
Sbjct 501 RLESLLQQLHASSSSSGKEQIKAACSDLEKIRKLKKEAEF 540
> At1g70070
Length=1171
Score = 27.7 bits (60), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 12/40 (30%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 4 LTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLA 43
+ WA G S+ ++ ++ EG + R +RR +LL Q+
Sbjct 1102 MVEAWASGLSWKEMMMECAMDEGDLARLLRRTIDLLAQIP 1141
> Hs5803029
Length=425
Score = 26.9 bits (58), Expect = 9.2, Method: Composition-based stats.
Identities = 14/48 (29%), Positives = 23/48 (47%), Gaps = 3/48 (6%)
Query 5 TYGWAQGQSFAALIRSTSLYEGTVIRCM---RRLEELLRQLASAAKAI 49
T GW +G + AL+ + S +E V+ + R EL R L + +
Sbjct 230 TCGWVKGSGYQALVHAASAFEVDVVVVLDQERLYNELKRDLPHFVRTV 277
> CE21767
Length=823
Score = 26.9 bits (58), Expect = 9.9, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 23/37 (62%), Gaps = 2/37 (5%)
Query 42 LASAAKAIGNQELEAKCVAAVRQLR--RGSIFSSSLY 76
LAS A G + +EA C A ++QL+ +G + S +L+
Sbjct 746 LASGMSASGRKAVEAMCEAVLKQLKTAKGFVTSKALF 782
Lambda K H
0.323 0.132 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1175087368
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40