bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2777_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
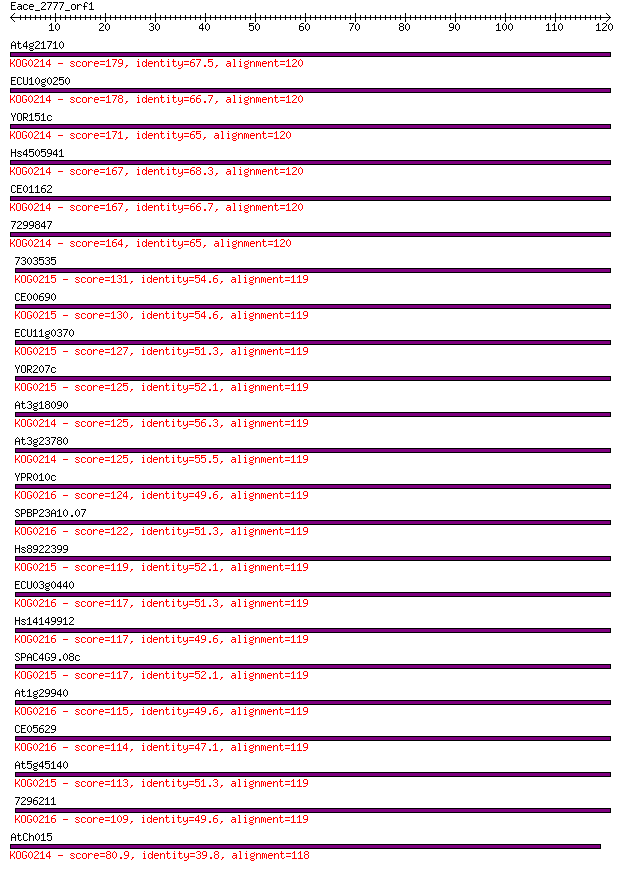
At4g21710 179 1e-45
ECU10g0250 178 2e-45
YOR151c 171 3e-43
Hs4505941 167 3e-42
CE01162 167 4e-42
7299847 164 5e-41
7303535 131 3e-31
CE00690 130 8e-31
ECU11g0370 127 5e-30
YOR207c 125 2e-29
At3g18090 125 2e-29
At3g23780 125 2e-29
YPR010c 124 4e-29
SPBP23A10.07 122 2e-28
Hs8922399 119 2e-27
ECU03g0440 117 4e-27
Hs14149912 117 5e-27
SPAC4G9.08c 117 5e-27
At1g29940 115 2e-26
CE05629 114 6e-26
At5g45140 113 8e-26
7296211 109 2e-24
AtCh015 80.9 5e-16
> At4g21710
Length=1188
Score = 179 bits (454), Expect = 1e-45, Method: Compositional matrix adjust.
Identities = 81/120 (67%), Positives = 99/120 (82%), Gaps = 0/120 (0%)
Query 1 VPSRMTIGHLVECLLGKTAAIIGGEGDATPFNDYTVSWIAGELHRLGFERHGNERLYHGH 60
+PSRMTIG L+EC++GK AA +G EGDATPF D TV I+ LH+ G++ G ER+Y+GH
Sbjct 978 IPSRMTIGQLIECIMGKVAAHMGKEGDATPFTDVTVDNISKALHKCGYQMRGFERMYNGH 1037
Query 61 TGLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKSREGGLRFGEMERD 120
TG + ++IF+GPTYYQRLKHMVDDKIH+R RGPV LTRQP EG+SR+GGLRFGEMERD
Sbjct 1038 TGRPLTAMIFLGPTYYQRLKHMVDDKIHSRGRGPVQILTRQPAEGRSRDGGLRFGEMERD 1097
> ECU10g0250
Length=1141
Score = 178 bits (451), Expect = 2e-45, Method: Composition-based stats.
Identities = 80/120 (66%), Positives = 97/120 (80%), Gaps = 0/120 (0%)
Query 1 VPSRMTIGHLVECLLGKTAAIIGGEGDATPFNDYTVSWIAGELHRLGFERHGNERLYHGH 60
+PSRMTIGHL+ECLLGK +A+ G EGDATPF+ TV I+ L GF++ G E +Y+G
Sbjct 941 IPSRMTIGHLIECLLGKVSAMSGEEGDATPFSGVTVDGISSRLKSYGFQQRGLEVMYNGM 1000
Query 61 TGLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKSREGGLRFGEMERD 120
TG + + +F GPTYYQRLKHMVDDKIHARARGP+ LTRQP+EG+SR+GGLRFGEMERD
Sbjct 1001 TGRKLRAQMFFGPTYYQRLKHMVDDKIHARARGPLQILTRQPVEGRSRDGGLRFGEMERD 1060
> YOR151c
Length=1224
Score = 171 bits (432), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 78/120 (65%), Positives = 95/120 (79%), Gaps = 0/120 (0%)
Query 1 VPSRMTIGHLVECLLGKTAAIIGGEGDATPFNDYTVSWIAGELHRLGFERHGNERLYHGH 60
+PSRMT+ HL+ECLL K AA+ G EGDA+PF D TV I+ L G++ G E +Y+GH
Sbjct 1017 IPSRMTVAHLIECLLSKVAALSGNEGDASPFTDITVEGISKLLREHGYQSRGFEVMYNGH 1076
Query 61 TGLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKSREGGLRFGEMERD 120
TG + + IF GPTYYQRL+HMVDDKIHARARGP+ LTRQP+EG+SR+GGLRFGEMERD
Sbjct 1077 TGKKLMAQIFFGPTYYQRLRHMVDDKIHARARGPMQVLTRQPVEGRSRDGGLRFGEMERD 1136
> Hs4505941
Length=1174
Score = 167 bits (424), Expect = 3e-42, Method: Compositional matrix adjust.
Identities = 82/121 (67%), Positives = 93/121 (76%), Gaps = 1/121 (0%)
Query 1 VPSRMTIGHLVECLLGKTAAIIGGEGDATPFND-YTVSWIAGELHRLGFERHGNERLYHG 59
+PSRMTIGHL+ECL GK +A G GDATPFND V I+ L G+ GNE LY+G
Sbjct 972 IPSRMTIGHLIECLQGKVSANKGEIGDATPFNDAVNVQKISNLLSDYGYHLRGNEVLYNG 1031
Query 60 HTGLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKSREGGLRFGEMER 119
TG + S IFIGPTYYQRLKHMVDDKIH+RARGP+ L RQPMEG+SR+GGLRFGEMER
Sbjct 1032 FTGRKITSQIFIGPTYYQRLKHMVDDKIHSRARGPIQILNRQPMEGRSRDGGLRFGEMER 1091
Query 120 D 120
D
Sbjct 1092 D 1092
> CE01162
Length=1193
Score = 167 bits (423), Expect = 4e-42, Method: Compositional matrix adjust.
Identities = 80/121 (66%), Positives = 94/121 (77%), Gaps = 1/121 (0%)
Query 1 VPSRMTIGHLVECLLGKTAAIIGGEGDATPFND-YTVSWIAGELHRLGFERHGNERLYHG 59
VPSRMTIGHL+ECL GK +A G GDATPFND V I+G L G+ GNE +Y+G
Sbjct 978 VPSRMTIGHLIECLQGKLSANKGEIGDATPFNDTVNVQKISGLLCEYGYHLRGNEVMYNG 1037
Query 60 HTGLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKSREGGLRFGEMER 119
HTG + + IF GPTYYQRLKHMVDDKIH+RARGP+ + RQPMEG++R+GGLRFGEMER
Sbjct 1038 HTGKKLTTQIFFGPTYYQRLKHMVDDKIHSRARGPIQMMNRQPMEGRARDGGLRFGEMER 1097
Query 120 D 120
D
Sbjct 1098 D 1098
> 7299847
Length=1176
Score = 164 bits (414), Expect = 5e-41, Method: Compositional matrix adjust.
Identities = 78/121 (64%), Positives = 93/121 (76%), Gaps = 1/121 (0%)
Query 1 VPSRMTIGHLVECLLGKTAAIIGGEGDATPFND-YTVSWIAGELHRLGFERHGNERLYHG 59
+PSRMTIGHL+ECL GK + G GDATPFND V I+ L G+ GNE +Y+G
Sbjct 974 IPSRMTIGHLIECLQGKLGSNKGEIGDATPFNDAVNVQKISTFLQEYGYHLRGNEVMYNG 1033
Query 60 HTGLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKSREGGLRFGEMER 119
HTG + + +F+GPTYYQRLKHMVDDKIH+RARGPV L RQPMEG++R+GGLRFGEMER
Sbjct 1034 HTGRKINAQVFLGPTYYQRLKHMVDDKIHSRARGPVQILVRQPMEGRARDGGLRFGEMER 1093
Query 120 D 120
D
Sbjct 1094 D 1094
> 7303535
Length=1137
Score = 131 bits (330), Expect = 3e-31, Method: Composition-based stats.
Identities = 65/119 (54%), Positives = 76/119 (63%), Gaps = 0/119 (0%)
Query 2 PSRMTIGHLVECLLGKTAAIIGGEGDATPFNDYTVSWIAGELHRLGFERHGNERLYHGHT 61
PSRMT+G +E L GK + G T F V I EL R GF G + Y G T
Sbjct 941 PSRMTVGKTLELLGGKAGLLEGKFHYGTAFGGSKVEDIQAELERHGFNYVGKDFFYSGIT 1000
Query 62 GLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKSREGGLRFGEMERD 120
G + + I+ GP YYQ+LKHMV DK+HARARGP A LTRQP +G+SREGGLR GEMERD
Sbjct 1001 GTPLEAYIYSGPVYYQKLKHMVQDKMHARARGPKAVLTRQPTQGRSREGGLRLGEMERD 1059
> CE00690
Length=1207
Score = 130 bits (326), Expect = 8e-31, Method: Composition-based stats.
Identities = 65/119 (54%), Positives = 77/119 (64%), Gaps = 0/119 (0%)
Query 2 PSRMTIGHLVECLLGKTAAIIGGEGDATPFNDYTVSWIAGELHRLGFERHGNERLYHGHT 61
PSRMT+G L+E L GK + G T F V + EL G+ G + L G T
Sbjct 1009 PSRMTVGKLMELLSGKAGVVNGTYHYGTAFGGDQVKDVCEELAACGYNYMGKDMLTSGIT 1068
Query 62 GLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKSREGGLRFGEMERD 120
G + + I+ GP YYQ+LKHMV DK+HARARGP AALTRQP EG+SREGGLR GEMERD
Sbjct 1069 GQPLSAYIYFGPIYYQKLKHMVLDKMHARARGPRAALTRQPTEGRSREGGLRLGEMERD 1127
> ECU11g0370
Length=1110
Score = 127 bits (319), Expect = 5e-30, Method: Composition-based stats.
Identities = 61/119 (51%), Positives = 77/119 (64%), Gaps = 0/119 (0%)
Query 2 PSRMTIGHLVECLLGKTAAIIGGEGDATPFNDYTVSWIAGELHRLGFERHGNERLYHGHT 61
PSRMT+G +VE + GK + G D+T F + +V L + GF G + G T
Sbjct 917 PSRMTVGKIVELISGKAGVLEGQILDSTAFKENSVEQTCELLIKHGFSYSGKDCFTSGTT 976
Query 62 GLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKSREGGLRFGEMERD 120
G + + IF GP +YQRLKHMV DKIH RARGP A LTRQP EG+S++GGL+ GEMERD
Sbjct 977 GAPLAAYIFFGPVFYQRLKHMVADKIHMRARGPRAILTRQPTEGRSKDGGLKLGEMERD 1035
> YOR207c
Length=1149
Score = 125 bits (314), Expect = 2e-29, Method: Composition-based stats.
Identities = 62/119 (52%), Positives = 77/119 (64%), Gaps = 0/119 (0%)
Query 2 PSRMTIGHLVECLLGKTAAIIGGEGDATPFNDYTVSWIAGELHRLGFERHGNERLYHGHT 61
PSRMT+G ++E + GK + G T F + ++ L GF G + LY G T
Sbjct 950 PSRMTVGKMIELISGKAGVLNGTLEYGTCFGGSKLEDMSKILVDQGFNYSGKDMLYSGIT 1009
Query 62 GLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKSREGGLRFGEMERD 120
G + + IF GP YYQ+LKHMV DK+HARARGP A LTRQP EG+SR+GGLR GEMERD
Sbjct 1010 GECLQAYIFFGPIYYQKLKHMVLDKMHARARGPRAVLTRQPTEGRSRDGGLRLGEMERD 1068
> At3g18090
Length=1038
Score = 125 bits (314), Expect = 2e-29, Method: Composition-based stats.
Identities = 67/131 (51%), Positives = 82/131 (62%), Gaps = 12/131 (9%)
Query 2 PSRMTIGHLVECLLGKTAA--IIGGEGD----------ATPFNDYTVSWIAGELHRLGFE 49
PSR T G L+E L K A I EG ATPF+ V+ I +LHR GF
Sbjct 817 PSRQTPGQLLEAALSKGIACPIQKKEGSSAAYTKLTRHATPFSTPGVTEITEQLHRAGFS 876
Query 50 RHGNERLYHGHTGLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKSRE 109
R GNER+Y+G +G M SLIF+GPT+YQRL HM ++K+ R GPV LTRQP+ + R
Sbjct 877 RWGNERVYNGRSGEMMRSLIFMGPTFYQRLVHMSENKVKFRNTGPVHPLTRQPVADRKRF 936
Query 110 GGLRFGEMERD 120
GG+RFGEMERD
Sbjct 937 GGIRFGEMERD 947
> At3g23780
Length=946
Score = 125 bits (314), Expect = 2e-29, Method: Composition-based stats.
Identities = 66/130 (50%), Positives = 82/130 (63%), Gaps = 11/130 (8%)
Query 2 PSRMTIGHLVECLLGK-TAAIIGGEGD----------ATPFNDYTVSWIAGELHRLGFER 50
PSR T G L+E L K A I EG ATPF+ V+ I +LHR GF R
Sbjct 726 PSRQTPGQLLEAALSKGIACPIQKEGSSAAYTKLTRHATPFSTPGVTEITEQLHRAGFSR 785
Query 51 HGNERLYHGHTGLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKSREG 110
GNER+Y+G +G M S+IF+GPT+YQRL HM +DK+ R GPV LTRQP+ + R G
Sbjct 786 WGNERVYNGRSGEMMRSMIFMGPTFYQRLVHMSEDKVKFRNTGPVHPLTRQPVADRKRFG 845
Query 111 GLRFGEMERD 120
G++FGEMERD
Sbjct 846 GIKFGEMERD 855
> YPR010c
Length=1203
Score = 124 bits (311), Expect = 4e-29, Method: Composition-based stats.
Identities = 59/123 (47%), Positives = 80/123 (65%), Gaps = 4/123 (3%)
Query 2 PSRMTIGHLVECLLGKTAAIIGGEGDATPF----NDYTVSWIAGELHRLGFERHGNERLY 57
PSRMTIG VE L GK A+ G D+TP+ +D + +L + G+ HGNE +Y
Sbjct 955 PSRMTIGMFVESLAGKAGALHGIAQDSTPWIFNEDDTPADYFGEQLAKAGYNYHGNEPMY 1014
Query 58 HGHTGLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKSREGGLRFGEM 117
G TG + + I++G YYQRL+HMV+DK R+ GPV +LT QP++G+ R GG+R GEM
Sbjct 1015 SGATGEELRADIYVGVVYYQRLRHMVNDKFQVRSTGPVNSLTMQPVKGRKRHGGIRVGEM 1074
Query 118 ERD 120
ERD
Sbjct 1075 ERD 1077
> SPBP23A10.07
Length=1227
Score = 122 bits (306), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 61/123 (49%), Positives = 76/123 (61%), Gaps = 4/123 (3%)
Query 2 PSRMTIGHLVECLLGKTAAIIGGEGDATPF----NDYTVSWIAGELHRLGFERHGNERLY 57
PSRMTIG +E L GK A G D+TPF + +L + G+ HGNE +Y
Sbjct 993 PSRMTIGMFIESLAGKAGACHGLAQDSTPFIYSEQQTAADYFGEQLVKAGYNYHGNEPMY 1052
Query 58 HGHTGLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKSREGGLRFGEM 117
G TG M + I+IG YYQRL+HMV DK R GP+ LTRQP++G+ R GG+RFGEM
Sbjct 1053 SGITGQEMKADIYIGVVYYQRLRHMVSDKFQVRTTGPIHNLTRQPVKGRKRAGGIRFGEM 1112
Query 118 ERD 120
ERD
Sbjct 1113 ERD 1115
> Hs8922399
Length=343
Score = 119 bits (297), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 62/119 (52%), Positives = 77/119 (64%), Gaps = 0/119 (0%)
Query 2 PSRMTIGHLVECLLGKTAAIIGGEGDATPFNDYTVSWIAGELHRLGFERHGNERLYHGHT 61
PSRMT+G L+E L GK + G T F V + +L R G+ G + + G T
Sbjct 145 PSRMTVGKLIELLAGKAGVLDGRFHYGTAFGGSKVKDVCEDLVRHGYNYLGKDYVTSGIT 204
Query 62 GLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKSREGGLRFGEMERD 120
G + + I+ GP YYQ+LKHMV DK+HARARGP A LTRQP EG+SR+GGLR GEMERD
Sbjct 205 GEPLEAYIYFGPVYYQKLKHMVLDKMHARARGPRAVLTRQPTEGRSRDGGLRLGEMERD 263
> ECU03g0440
Length=1062
Score = 117 bits (294), Expect = 4e-27, Method: Composition-based stats.
Identities = 61/133 (45%), Positives = 77/133 (57%), Gaps = 14/133 (10%)
Query 2 PSRMTIGHLVECLLGKTAAIIGGEGDATPF-----------NDYTV---SWIAGELHRLG 47
PSRMTIG L+E + GK + G E D T F +D V ++ EL R G
Sbjct 855 PSRMTIGMLIESIAGKVGCLSGNEQDGTVFKKSFLLEQEEGDDEKVRRKEYLCSELRRHG 914
Query 48 FERHGNERLYHGHTGLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKS 107
F +GNE +Y G G + IF+G YYQRLKHMV DK R +G V + TRQP+ G+
Sbjct 915 FNYYGNEPMYSGVAGNEFRADIFVGVVYYQRLKHMVGDKFQVRTKGAVVSTTRQPVGGRK 974
Query 108 REGGLRFGEMERD 120
+ GG+RFGEMERD
Sbjct 975 KRGGIRFGEMERD 987
> Hs14149912
Length=459
Score = 117 bits (293), Expect = 5e-27, Method: Composition-based stats.
Identities = 59/123 (47%), Positives = 78/123 (63%), Gaps = 4/123 (3%)
Query 2 PSRMTIGHLVECLLGKTAAIIGGEGDATPF----NDYTVSWIAGELHRLGFERHGNERLY 57
PSRMTIG L+E + GK+AA+ G DATPF + + + L G+ +G ERLY
Sbjct 280 PSRMTIGMLIESMAGKSAALHGLCHDATPFIFSEENSALEYFGEMLKAAGYNFYGTERLY 339
Query 58 HGHTGLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKSREGGLRFGEM 117
G +GL + + IFIG YYQRL+HMV DK R G +T QP+ G++ +GG+RFGEM
Sbjct 340 SGISGLELEADIFIGVVYYQRLRHMVSDKFQVRTTGARDRVTNQPIGGRNVQGGIRFGEM 399
Query 118 ERD 120
ERD
Sbjct 400 ERD 402
> SPAC4G9.08c
Length=1165
Score = 117 bits (293), Expect = 5e-27, Method: Compositional matrix adjust.
Identities = 62/119 (52%), Positives = 76/119 (63%), Gaps = 0/119 (0%)
Query 2 PSRMTIGHLVECLLGKTAAIIGGEGDATPFNDYTVSWIAGELHRLGFERHGNERLYHGHT 61
PSRMT+G ++E L GK + G T F V + L G+ G + L G T
Sbjct 966 PSRMTVGKMIELLSGKVGVLRGTLEYGTCFGGTKVEDASRILVEHGYNYSGKDMLTSGIT 1025
Query 62 GLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKSREGGLRFGEMERD 120
G + + IF+GP YYQ+LKHMV DK+HARARGP A LTRQP EG+SR+GGLR GEMERD
Sbjct 1026 GETLEAYIFMGPIYYQKLKHMVMDKMHARARGPRAVLTRQPTEGRSRDGGLRLGEMERD 1084
> At1g29940
Length=1114
Score = 115 bits (288), Expect = 2e-26, Method: Composition-based stats.
Identities = 59/133 (44%), Positives = 75/133 (56%), Gaps = 14/133 (10%)
Query 2 PSRMTIGHLVECLLGKTAAIIGGEGDATPFNDYT--------------VSWIAGELHRLG 47
PSRMTI L+E + K ++ G DATPF D V + L G
Sbjct 874 PSRMTIAMLLESIAAKGGSLHGKFVDATPFRDAVKKTNGEEESKSSLLVDDLGSMLKEKG 933
Query 48 FERHGNERLYHGHTGLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKS 107
F +G E LY G+ G+ + IF+GP YYQRL+HMV DK R+ G V LT QP++G+
Sbjct 934 FNHYGTETLYSGYLGVELKCEIFMGPVYYQRLRHMVSDKFQVRSTGQVDQLTHQPIKGRK 993
Query 108 REGGLRFGEMERD 120
R GG+RFGEMERD
Sbjct 994 RGGGIRFGEMERD 1006
> CE05629
Length=1127
Score = 114 bits (284), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 56/123 (45%), Positives = 76/123 (61%), Gaps = 4/123 (3%)
Query 2 PSRMTIGHLVECLLGKTAAIIGGEGDATPF----NDYTVSWIAGELHRLGFERHGNERLY 57
PSRMTIG ++E + GK AA G DA+PF ++ ++ L + G+ +GNE Y
Sbjct 895 PSRMTIGMMIESMAGKAAATHGENYDASPFVFNEDNTAINHFGELLTKAGYNYYGNETFY 954
Query 58 HGHTGLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKSREGGLRFGEM 117
G G M IF G YYQRL+HM+ DK RA GP+ +T QP++G+ + GG+RFGEM
Sbjct 955 SGVDGRQMEMQIFFGIVYYQRLRHMIADKFQVRATGPIDPITHQPVKGRKKGGGIRFGEM 1014
Query 118 ERD 120
ERD
Sbjct 1015 ERD 1017
> At5g45140
Length=1194
Score = 113 bits (283), Expect = 8e-26, Method: Composition-based stats.
Identities = 61/126 (48%), Positives = 77/126 (61%), Gaps = 9/126 (7%)
Query 2 PSRMTIGHLVECLLGKTAAIIGGE-------GDATPFNDYTVSWIAGELHRLGFERHGNE 54
PSRMT+G ++E LLG A + G G+ + D V I+ L GF G +
Sbjct 964 PSRMTVGKMIE-LLGSKAGVSCGRFHYGSAFGERSGHAD-KVETISATLVEKGFSYSGKD 1021
Query 55 RLYHGHTGLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKSREGGLRF 114
LY G +G + + IF+GP YYQ+LKHMV DK+HAR GP +TRQP EGKS+ GGLR
Sbjct 1022 LLYSGISGEPVEAYIFMGPIYYQKLKHMVLDKMHARGSGPRVMMTRQPTEGKSKNGGLRV 1081
Query 115 GEMERD 120
GEMERD
Sbjct 1082 GEMERD 1087
> 7296211
Length=1129
Score = 109 bits (272), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 59/123 (47%), Positives = 75/123 (60%), Gaps = 4/123 (3%)
Query 2 PSRMTIGHLVECLLGKTAAIIGGEGDATPF----NDYTVSWIAGELHRLGFERHGNERLY 57
PSRMTI ++E + GK AAI G DATPF + + + L G+ +G ERLY
Sbjct 912 PSRMTIAMMIETMAGKGAAIHGNVYDATPFRFSEENTAIDYFGKMLEAGGYNYYGTERLY 971
Query 58 HGHTGLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEGKSREGGLRFGEM 117
G G M + IF G +YQRL+HMV DK R+ G V A T QP++G+ R GG+RFGEM
Sbjct 972 SGVDGREMTADIFFGVVHYQRLRHMVFDKWQVRSTGAVEARTHQPIKGRKRGGGVRFGEM 1031
Query 118 ERD 120
ERD
Sbjct 1032 ERD 1034
> AtCh015
Length=1072
Score = 80.9 bits (198), Expect = 5e-16, Method: Composition-based stats.
Identities = 47/133 (35%), Positives = 68/133 (51%), Gaps = 15/133 (11%)
Query 1 VPSRMTIGHLVECLLGKTAAIIGGEGDATPFNDY-----TVSWIAGELHRLG-------- 47
VPSRM +G + EC LG +++ PF++ + + EL+
Sbjct 859 VPSRMNVGQIFECSLGLAGSLLDRHYRIAPFDERYEQEASRKLVFSELYEASKQTANPWV 918
Query 48 FERH--GNERLYHGHTGLHMPSLIFIGPTYYQRLKHMVDDKIHARARGPVAALTRQPMEG 105
FE G R++ G TG + IG Y +L H VDDKIH R+ G A +T+QP+ G
Sbjct 919 FEPEYPGKSRIFDGRTGDPFEQPVIIGKPYILKLIHQVDDKIHGRSSGHYALVTQQPLRG 978
Query 106 KSREGGLRFGEME 118
+S++GG R GEME
Sbjct 979 RSKQGGQRVGEME 991
Lambda K H
0.322 0.141 0.440
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160968786
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40