bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2773_orf3
Length=93
Score E
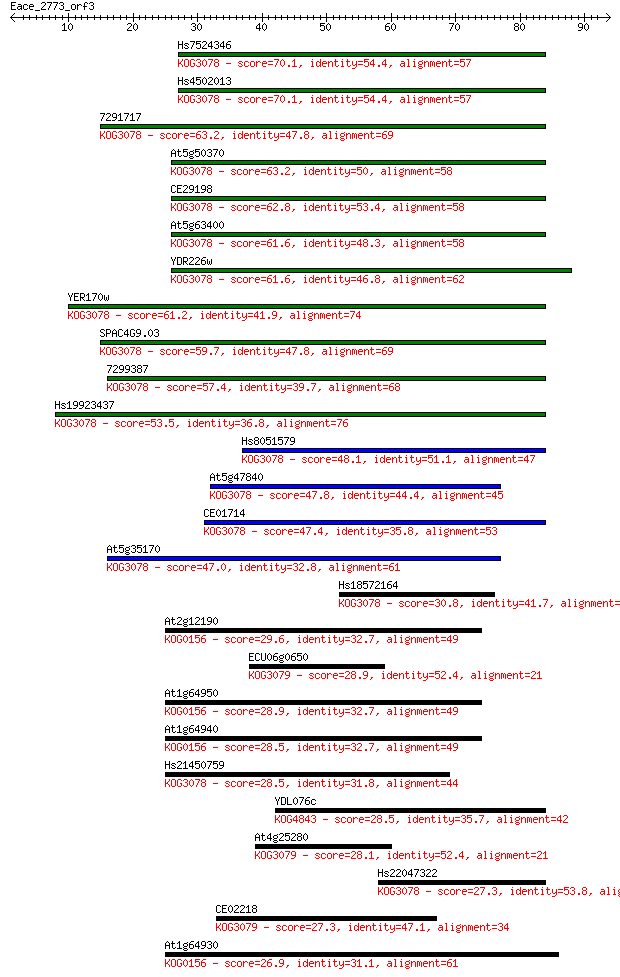
Sequences producing significant alignments: (Bits) Value
Hs7524346 70.1 8e-13
Hs4502013 70.1 8e-13
7291717 63.2 1e-10
At5g50370 63.2 1e-10
CE29198 62.8 1e-10
At5g63400 61.6 3e-10
YDR226w 61.6 3e-10
YER170w 61.2 4e-10
SPAC4G9.03 59.7 1e-09
7299387 57.4 6e-09
Hs19923437 53.5 9e-08
Hs8051579 48.1 4e-06
At5g47840 47.8 5e-06
CE01714 47.4 6e-06
At5g35170 47.0 8e-06
Hs18572164 30.8 0.67
At2g12190 29.6 1.3
ECU06g0650 28.9 2.3
At1g64950 28.9 2.5
At1g64940 28.5 2.9
Hs21450759 28.5 3.1
YDL076c 28.5 3.3
At4g25280 28.1 3.8
Hs22047322 27.3 6.7
CE02218 27.3 7.8
At1g64930 26.9 7.9
> Hs7524346
Length=232
Score = 70.1 bits (170), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 31/57 (54%), Positives = 39/57 (68%), Gaps = 0/57 (0%)
Query 27 LDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
LD L+ +K+KLD V+ F P+ L++RI GR IHP SGR YH F PPK P KDD+
Sbjct 111 LDDLMEKRKEKLDSVIEFSIPDSLLIRRITGRLIHPKSGRSYHEEFNPPKEPMKDDI 167
> Hs4502013
Length=239
Score = 70.1 bits (170), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 31/57 (54%), Positives = 39/57 (68%), Gaps = 0/57 (0%)
Query 27 LDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
LD L+ +K+KLD V+ F P+ L++RI GR IHP SGR YH F PPK P KDD+
Sbjct 111 LDDLMEKRKEKLDSVIEFSIPDSLLIRRITGRLIHPKSGRSYHEEFNPPKEPMKDDI 167
> 7291717
Length=237
Score = 63.2 bits (152), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 33/69 (47%), Positives = 40/69 (57%), Gaps = 0/69 (0%)
Query 15 RWFIFAFSLHFQLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRP 74
R + A + +LD LL +K LD V+ F + LV+RI GR IH ASGR YH F P
Sbjct 99 RTVVQAEKVATRLDTLLDKRKTNLDAVIEFAIDDSLLVRRITGRLIHQASGRSYHEEFAP 158
Query 75 PKVPGKDDV 83
PK P DDV
Sbjct 159 PKKPMTDDV 167
> At5g50370
Length=248
Score = 63.2 bits (152), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 29/58 (50%), Positives = 40/58 (68%), Gaps = 0/58 (0%)
Query 26 QLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
+LD++L + ++D V+ F + L +RI GR IHP+SGR YHT F PPKVPG DD+
Sbjct 129 KLDEMLNRRGAQIDKVLNFAIDDSVLEERITGRWIHPSSGRSYHTKFAPPKVPGVDDL 186
> CE29198
Length=251
Score = 62.8 bits (151), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 31/58 (53%), Positives = 41/58 (70%), Gaps = 0/58 (0%)
Query 26 QLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
+LD++L +K LD VV F+ ++ LV+RI GR H ASGR YH F+PPKVP KDD+
Sbjct 121 KLDEILERRKTPLDTVVEFNIADDLLVRRITGRLFHIASGRSYHLEFKPPKVPMKDDL 178
> At5g63400
Length=246
Score = 61.6 bits (148), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 28/58 (48%), Positives = 39/58 (67%), Gaps = 0/58 (0%)
Query 26 QLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
+LD++L + ++D V+ F + L +RI GR IHP+SGR YHT F PPK PG DD+
Sbjct 128 KLDEMLKRRGTEIDKVLNFAIDDAILEERITGRWIHPSSGRSYHTKFAPPKTPGVDDI 185
> YDR226w
Length=222
Score = 61.6 bits (148), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 29/62 (46%), Positives = 37/62 (59%), Gaps = 0/62 (0%)
Query 26 QLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDVRH 85
+LD++L Q L+ + +E LV RI GR IHPASGR YH +F PPK KDDV
Sbjct 102 KLDQMLKEQGTPLEKAIELKVDDELLVARITGRLIHPASGRSYHKIFNPPKEDMKDDVTG 161
Query 86 DC 87
+
Sbjct 162 EA 163
> YER170w
Length=225
Score = 61.2 bits (147), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 31/74 (41%), Positives = 42/74 (56%), Gaps = 0/74 (0%)
Query 10 ALRLFRWFIFAFSLHFQLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYH 69
A+ L F + LD+LL L+ VV D PE T+++RI R +H SGRVY+
Sbjct 97 AMWLLDGFPRTTAQASALDELLKQHDASLNLVVELDVPESTILERIENRYVHVPSGRVYN 156
Query 70 TVFRPPKVPGKDDV 83
+ PPKVPG DD+
Sbjct 157 LQYNPPKVPGLDDI 170
> SPAC4G9.03
Length=220
Score = 59.7 bits (143), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 33/69 (47%), Positives = 40/69 (57%), Gaps = 4/69 (5%)
Query 15 RWFIFAFSLHFQLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRP 74
R + A L LD+L K L+ V+ +E LV+RI GR +HP SGR YH F P
Sbjct 92 RTVVQAEKLTALLDEL----KLDLNTVLELQVDDELLVRRITGRLVHPGSGRSYHLEFNP 147
Query 75 PKVPGKDDV 83
PKVP KDDV
Sbjct 148 PKVPMKDDV 156
> 7299387
Length=216
Score = 57.4 bits (137), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 27/68 (39%), Positives = 41/68 (60%), Gaps = 1/68 (1%)
Query 16 WFIFAFSLHFQLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPP 75
+ + F + + LA+++Q +D V+ D P ++ R+ R IH SGRVY+ F+ P
Sbjct 85 YILDGFPRNIAQAEALAAREQ-IDAVITLDVPHSVIIDRVKNRWIHAPSGRVYNIGFKNP 143
Query 76 KVPGKDDV 83
KVPGKDDV
Sbjct 144 KVPGKDDV 151
> Hs19923437
Length=227
Score = 53.5 bits (127), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 28/76 (36%), Positives = 39/76 (51%), Gaps = 1/76 (1%)
Query 8 FQALRLFRWFIFAFSLHFQLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRV 67
+ L + W + F + L Q +D V+ + P E + QR+ R IHPASGRV
Sbjct 79 LKNLTQYSWLLDGFPRTLPQAEALDRAYQ-IDTVINLNVPFEVIKQRLTARWIHPASGRV 137
Query 68 YHTVFRPPKVPGKDDV 83
Y+ F PPK G DD+
Sbjct 138 YNIEFNPPKTVGIDDL 153
> Hs8051579
Length=223
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 24/47 (51%), Positives = 30/47 (63%), Gaps = 0/47 (0%)
Query 37 KLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
++D V+ + P ETL R+ R IHP SGRVY+ F PP V G DDV
Sbjct 105 EVDLVISLNIPFETLKDRLSRRWIHPPSGRVYNLDFNPPHVHGIDDV 151
> At5g47840
Length=283
Score = 47.8 bits (112), Expect = 5e-06, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 30/52 (57%), Gaps = 7/52 (13%)
Query 32 ASQKQKLDGVVF-------FDTPEETLVQRICGRRIHPASGRVYHTVFRPPK 76
ASQ L G F + PEE L++R+ GRR+ P +G++YH + PP+
Sbjct 155 ASQATALKGFGFQPDLFIVLEVPEEILIERVVGRRLDPVTGKIYHLKYSPPE 206
> CE01714
Length=222
Score = 47.4 bits (111), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 31/53 (58%), Gaps = 0/53 (0%)
Query 31 LASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
+ ++ L+ +V P + L+ R+ + +HPASGR Y+ PPK GKDD+
Sbjct 97 MVEEQAPLNLIVELKVPRKVLIDRLSKQLVHPASGRAYNLEVNPPKEEGKDDI 149
> At5g35170
Length=498
Score = 47.0 bits (110), Expect = 8e-06, Method: Composition-based stats.
Identities = 20/61 (32%), Positives = 30/61 (49%), Gaps = 0/61 (0%)
Query 16 WFIFAFSLHFQLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPP 75
W + F F + L K D + D P+E L+ R GRR+ P +G++YH PP
Sbjct 79 WLLDGFPRSFAQAQSLDKLNVKPDIFILLDVPDEILIDRCVGRRLDPVTGKIYHIKNYPP 138
Query 76 K 76
+
Sbjct 139 E 139
> Hs18572164
Length=94
Score = 30.8 bits (68), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 10/24 (41%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 52 VQRICGRRIHPASGRVYHTVFRPP 75
++R+ RRI P +G YH +++PP
Sbjct 1 MERLTLRRIDPVTGERYHLMYKPP 24
> At2g12190
Length=512
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 22/49 (44%), Gaps = 0/49 (0%)
Query 25 FQLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFR 73
F D+ LA Q L+G VF D P + +I H S +Y +R
Sbjct 81 FVADRSLAHQALVLNGAVFADRPPAAPISKIISSNQHNISSSLYGATWR 129
> ECU06g0650
Length=123
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 11/21 (52%), Positives = 15/21 (71%), Gaps = 0/21 (0%)
Query 38 LDGVVFFDTPEETLVQRICGR 58
+D V+F D E+T + RICGR
Sbjct 46 VDLVIFIDVDEDTCISRICGR 66
> At1g64950
Length=510
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 22/49 (44%), Gaps = 0/49 (0%)
Query 25 FQLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFR 73
F D+ LA Q L+G VF D P + +I H S +Y +R
Sbjct 81 FVADRSLAHQALVLNGAVFADRPPAAPISKIISSNQHNISSCLYGATWR 129
> At1g64940
Length=511
Score = 28.5 bits (62), Expect = 2.9, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 22/49 (44%), Gaps = 0/49 (0%)
Query 25 FQLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFR 73
F D+ LA Q L+G VF D P + +I H S +Y +R
Sbjct 82 FVTDRSLAHQALVLNGAVFADRPPAESISKIISSNQHNISSCLYGATWR 130
> Hs21450759
Length=421
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 14/44 (31%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 25 FQLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVY 68
Q +L+ + K D ++ P+ L QRI G+R H +G +Y
Sbjct 128 LQQIELIKNLNLKPDVIINIKCPDYDLCQRISGQRQHNNTGYIY 171
> YDL076c
Length=294
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 15/43 (34%), Positives = 23/43 (53%), Gaps = 1/43 (2%)
Query 42 VFFDTPEETLVQRICGRRIHPASGRVYHTVFRPP-KVPGKDDV 83
++ D + LV R CG +I SG +H + R P V +D+V
Sbjct 170 IYSDDSDPILVLRHCGFKIGAPSGGSFHKLRRTPVNVTNQDNV 212
> At4g25280
Length=227
Score = 28.1 bits (61), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 11/21 (52%), Positives = 15/21 (71%), Gaps = 0/21 (0%)
Query 39 DGVVFFDTPEETLVQRICGRR 59
D V+FFD PEE +V+R+ R
Sbjct 147 DVVLFFDCPEEEMVKRVLNRN 167
> Hs22047322
Length=225
Score = 27.3 bits (59), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 14/27 (51%), Positives = 17/27 (62%), Gaps = 1/27 (3%)
Query 58 RRIHP-ASGRVYHTVFRPPKVPGKDDV 83
R +HP SG+VY F PPK G DD+
Sbjct 125 RFLHPDNSGQVYGIEFNPPKTVGIDDL 151
> CE02218
Length=210
Score = 27.3 bits (59), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 21/34 (61%), Gaps = 2/34 (5%)
Query 33 SQKQKLDGVVFFDTPEETLVQRICGRRIHPASGR 66
S+ Q+ V+FFD EETLV+R+ R SGR
Sbjct 120 SEIQEAKLVLFFDVAEETLVKRLLHR--AQTSGR 151
> At1g64930
Length=511
Score = 26.9 bits (58), Expect = 7.9, Method: Composition-based stats.
Identities = 19/72 (26%), Positives = 27/72 (37%), Gaps = 11/72 (15%)
Query 25 FQLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFR----------- 73
F D LA Q L+G VF D P + +I H + +Y +R
Sbjct 81 FVADGSLAHQALVLNGAVFADRPPAAPISKILSNNQHTITSCLYGVTWRLLRRNITEILH 140
Query 74 PPKVPGKDDVRH 85
P ++ VRH
Sbjct 141 PSRMKSYSHVRH 152
Lambda K H
0.332 0.147 0.480
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167934574
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40