bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2730_orf1
Length=177
Score E
Sequences producing significant alignments: (Bits) Value
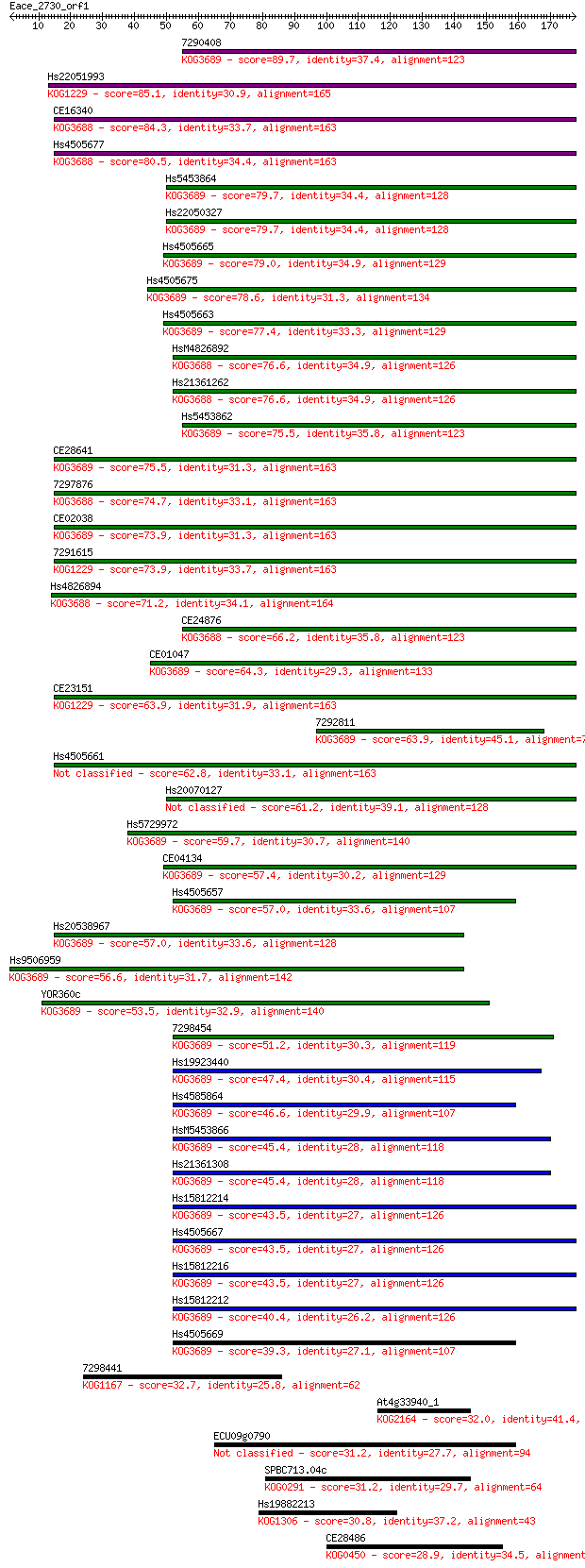
7290408 89.7 3e-18
Hs22051993 85.1 7e-17
CE16340 84.3 1e-16
Hs4505677 80.5 2e-15
Hs5453864 79.7 3e-15
Hs22050327 79.7 3e-15
Hs4505665 79.0 4e-15
Hs4505675 78.6 5e-15
Hs4505663 77.4 1e-14
HsM4826892 76.6 3e-14
Hs21361262 76.6 3e-14
Hs5453862 75.5 5e-14
CE28641 75.5 5e-14
7297876 74.7 8e-14
CE02038 73.9 1e-13
7291615 73.9 2e-13
Hs4826894 71.2 9e-13
CE24876 66.2 3e-11
CE01047 64.3 1e-10
CE23151 63.9 1e-10
7292811 63.9 2e-10
Hs4505661 62.8 4e-10
Hs20070127 61.2 1e-09
Hs5729972 59.7 3e-09
CE04134 57.4 2e-08
Hs4505657 57.0 2e-08
Hs20538967 57.0 2e-08
Hs9506959 56.6 2e-08
YOR360c 53.5 2e-07
7298454 51.2 9e-07
Hs19923440 47.4 1e-05
Hs4585864 46.6 3e-05
HsM5453866 45.4 5e-05
Hs21361308 45.4 5e-05
Hs15812214 43.5 2e-04
Hs4505667 43.5 2e-04
Hs15812216 43.5 2e-04
Hs15812212 40.4 0.002
Hs4505669 39.3 0.004
7298441 32.7 0.34
At4g33940_1 32.0 0.62
ECU09g0790 31.2 1.2
SPBC713.04c 31.2 1.3
Hs19882213 30.8 1.4
CE28486 28.9 6.3
> 7290408
Length=624
Score = 89.7 bits (221), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 46/124 (37%), Positives = 68/124 (54%), Gaps = 1/124 (0%)
Query 55 FLQTLQWQY-QPNPYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHDVGH 113
F+ TL+ Y + NP+HN LHAA V + N + + PLE A HDV H
Sbjct 271 FMSTLEDHYVKDNPFHNSLHAADVTQSTNVLLNTPALEGVFTPLEVGGALFAACIHDVDH 330
Query 114 PGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRILERKSCNIFFLLSVGAFRSVRAKI 173
PG TNQF V LA++YND S LEN H + F++L+ + C+IF + +++R +
Sbjct 331 PGLTNQFLVNSSSELALMYNDESVLENHHLAVAFKLLQNQGCDIFCNMQKKQRQTLRKMV 390
Query 174 IECI 177
I+ +
Sbjct 391 IDIV 394
> Hs22051993
Length=761
Score = 85.1 bits (209), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 51/170 (30%), Positives = 82/170 (48%), Gaps = 5/170 (2%)
Query 13 KQWSFSVLQLDQEVNG-ALFVVGTALLGP--VATDWGCGSNELSQFLQTLQWQYQP-NPY 68
+ W F++ +L+ + L +G + V C L + Q ++ Y N Y
Sbjct 431 ESWDFNIFELEAITHKRPLVYLGLKVFSRFGVCEFLNCSETTLRAWFQVIEANYHSSNAY 490
Query 69 HNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHDVGHPGRTNQFFVACFDPL 128
HN HAA V H V ++ L++ A +A HDV HPGRTN F L
Sbjct 491 HNSTHAADVLHATAFFLGKERVKGSLDQLDEVAALIAATVHDVDHPGRTNSFLCNAGSEL 550
Query 129 AIIYNDISPLENFHSCLCFRI-LERKSCNIFFLLSVGAFRSVRAKIIECI 177
A++YND + LE+ H+ L F++ ++ CNIF + +R++R II+ +
Sbjct 551 AVLYNDTAVLESHHTALAFQLTVKDTKCNIFKNIDRNHYRTLRQAIIDMV 600
> CE16340
Length=664
Score = 84.3 bits (207), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 55/168 (32%), Positives = 79/168 (47%), Gaps = 5/168 (2%)
Query 15 WSFSVLQLDQEVNG-ALFVVGTALLGPVA--TDWGCGSNELSQFLQTLQWQYQP--NPYH 69
WSFS QL++ G AL VG L + L +L L+ Y NPYH
Sbjct 274 WSFSPFQLNEVSEGHALKYVGFELFNRYGFMDRFKVPLTALENYLSALEVGYSKHNNPYH 333
Query 70 NQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHDVGHPGRTNQFFVACFDPLA 129
N +HAA V + + + G+ + LE A+ L HD H G TN F + A
Sbjct 334 NVVHAADVTQSSHFMLSQTGLANSLGDLELLAVLFGALIHDYEHTGHTNNFHIQSQSQFA 393
Query 130 IIYNDISPLENFHSCLCFRILERKSCNIFFLLSVGAFRSVRAKIIECI 177
++YND S LEN H CFR+++ NI L+ ++ +R +IE +
Sbjct 394 MLYNDRSVLENHHVSSCFRLMKEDDKNILTHLTRDEYKELRNMVIEIV 441
> Hs4505677
Length=536
Score = 80.5 bits (197), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 56/168 (33%), Positives = 78/168 (46%), Gaps = 5/168 (2%)
Query 15 WSFSVLQLDQEVNG-ALFVVGTALLGP--VATDWGCGSNELSQFLQTLQWQY--QPNPYH 69
W F V L+Q + AL + LL + + + + L FL L+ Y NPYH
Sbjct 164 WCFDVFSLNQAADDHALRTIVFELLTRHNLISRFKIPTVFLMSFLDALETGYGKYKNPYH 223
Query 70 NQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHDVGHPGRTNQFFVACFDPLA 129
NQ+HAA V G+ + +E A+ A HD H G TN F + A
Sbjct 224 NQIHAADVTQTVHCFLLRTGMVHCLSEIELLAIIFAAAIHDYEHTGTTNSFHIQTKSECA 283
Query 130 IIYNDISPLENFHSCLCFRILERKSCNIFFLLSVGAFRSVRAKIIECI 177
I+YND S LEN H FR+++ NIF L+ F +RA +IE +
Sbjct 284 IVYNDRSVLENHHISSVFRLMQDDEMNIFINLTKDEFVELRALVIEMV 331
> Hs5453864
Length=809
Score = 79.7 bits (195), Expect = 3e-15, Method: Composition-based stats.
Identities = 44/129 (34%), Positives = 71/129 (55%), Gaps = 1/129 (0%)
Query 50 NELSQFLQTLQWQYQPN-PYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLA 108
+ L +L TL+ Y + YHN +HAA V + + + + LE A A+
Sbjct 442 DTLITYLMTLEDHYHADVAYHNNIHAADVVQSTHVLLSTPALEAVFTDLEILAAIFASAI 501
Query 109 HDVGHPGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRILERKSCNIFFLLSVGAFRS 168
HDV HPG +NQF + LA++YND S LEN H + F++L+ ++C+IF L+ +S
Sbjct 502 HDVDHPGVSNQFLINTNSELALMYNDSSVLENHHLAVGFKLLQEENCDIFQNLTKKQRQS 561
Query 169 VRAKIIECI 177
+R +I+ +
Sbjct 562 LRKMVIDIV 570
> Hs22050327
Length=641
Score = 79.7 bits (195), Expect = 3e-15, Method: Composition-based stats.
Identities = 44/129 (34%), Positives = 71/129 (55%), Gaps = 1/129 (0%)
Query 50 NELSQFLQTLQWQYQPN-PYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLA 108
+ L +L TL+ Y + YHN +HAA V + + + + LE A A+
Sbjct 274 DTLITYLMTLEDHYHADVAYHNNIHAADVVQSTHVLLSTPALEAVFTDLEILAAIFASAI 333
Query 109 HDVGHPGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRILERKSCNIFFLLSVGAFRS 168
HDV HPG +NQF + LA++YND S LEN H + F++L+ ++C+IF L+ +S
Sbjct 334 HDVDHPGVSNQFLINTNSELALMYNDSSVLENHHLAVGFKLLQEENCDIFQNLTKKQRQS 393
Query 169 VRAKIIECI 177
+R +I+ +
Sbjct 394 LRKMVIDIV 402
> Hs4505665
Length=712
Score = 79.0 bits (193), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 45/130 (34%), Positives = 70/130 (53%), Gaps = 1/130 (0%)
Query 49 SNELSQFLQTLQWQYQPN-PYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATL 107
++ L+ +L L+ Y N YHN LHAA V + + + LE A A+
Sbjct 367 ADTLATYLLMLEGHYHANVAYHNSLHAADVAQSTHVLLATPALEAVFTDLEILAALFASA 426
Query 108 AHDVGHPGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRILERKSCNIFFLLSVGAFR 167
HDV HPG +NQF + +A++YND S LEN H + F++L+ ++C+IF LS
Sbjct 427 IHDVDHPGVSNQFLINTNSDVALMYNDASVLENHHLAVGFKLLQAENCDIFQNLSAKQRL 486
Query 168 SVRAKIIECI 177
S+R +I+ +
Sbjct 487 SLRRMVIDMV 496
> Hs4505675
Length=593
Score = 78.6 bits (192), Expect = 5e-15, Method: Composition-based stats.
Identities = 42/134 (31%), Positives = 64/134 (47%), Gaps = 0/134 (0%)
Query 44 DWGCGSNELSQFLQTLQWQYQPNPYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALA 103
D+ L ++L + Y+ NP+HN H V S+ + + + + L
Sbjct 287 DFSINPVTLRRWLFCVHDNYRNNPFHNFRHCFCVAQMMYSMVWLCSLQEKFSQTDILILM 346
Query 104 VATLAHDVGHPGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRILERKSCNIFFLLSV 163
A + HD+ HPG N + + LA+ YNDISPLEN H + F+IL CNIF +
Sbjct 347 TAAICHDLDHPGYNNTYQINARTELAVRYNDISPLENHHCAVAFQILAEPECNIFSNIPP 406
Query 164 GAFRSVRAKIIECI 177
F+ +R +I I
Sbjct 407 DGFKQIRQGMITLI 420
> Hs4505663
Length=564
Score = 77.4 bits (189), Expect = 1e-14, Method: Composition-based stats.
Identities = 43/130 (33%), Positives = 69/130 (53%), Gaps = 1/130 (0%)
Query 49 SNELSQFLQTLQWQYQPN-PYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATL 107
S+ ++ TL+ Y + YHN LHAA V + + + + LE A A
Sbjct 213 SDTFITYMMTLEDHYHSDVAYHNSLHAADVAQSTHVLLSTPALDAVFTDLEILAAIFAAA 272
Query 108 AHDVGHPGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRILERKSCNIFFLLSVGAFR 167
HDV HPG +NQF + LA++YND S LEN H + F++L+ + C+IF L+ +
Sbjct 273 IHDVDHPGVSNQFLINTNSELALMYNDESVLENHHLAVGFKLLQEEHCDIFMNLTKKQRQ 332
Query 168 SVRAKIIECI 177
++R +I+ +
Sbjct 333 TLRKMVIDMV 342
> HsM4826892
Length=535
Score = 76.6 bits (187), Expect = 3e-14, Method: Composition-based stats.
Identities = 44/128 (34%), Positives = 63/128 (49%), Gaps = 2/128 (1%)
Query 52 LSQFLQTLQWQYQP--NPYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAH 109
L F + L+ Y NPYHN +HAA V + G+ + LE A+ A H
Sbjct 200 LITFAEALEVGYSKYKNPYHNLIHAADVTQTVHYIMLHTGIMHWLTELEILAMVFAAAIH 259
Query 110 DVGHPGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRILERKSCNIFFLLSVGAFRSV 169
D H G TN F + +AI+YND S LEN H +R+++ + NI LS +R +
Sbjct 260 DYEHTGTTNNFHIQTRSDVAILYNDRSVLENHHVSAAYRLMQEEEMNILINLSKDDWRDL 319
Query 170 RAKIIECI 177
R +IE +
Sbjct 320 RNLVIEMV 327
> Hs21361262
Length=545
Score = 76.6 bits (187), Expect = 3e-14, Method: Composition-based stats.
Identities = 44/128 (34%), Positives = 63/128 (49%), Gaps = 2/128 (1%)
Query 52 LSQFLQTLQWQYQP--NPYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAH 109
L F + L+ Y NPYHN +HAA V + G+ + LE A+ A H
Sbjct 200 LITFAEALEVGYSKYKNPYHNLIHAADVTQTVHYIMLHTGIMHWLTELEILAMVFAAAIH 259
Query 110 DVGHPGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRILERKSCNIFFLLSVGAFRSV 169
D H G TN F + +AI+YND S LEN H +R+++ + NI LS +R +
Sbjct 260 DYEHTGTTNNFHIQTRSDVAILYNDRSVLENHHVSAAYRLMQEEEMNILINLSKDDWRDL 319
Query 170 RAKIIECI 177
R +IE +
Sbjct 320 RNLVIEMV 327
> Hs5453862
Length=647
Score = 75.5 bits (184), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 44/124 (35%), Positives = 66/124 (53%), Gaps = 1/124 (0%)
Query 55 FLQTLQWQYQPN-PYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHDVGH 113
++ TL+ Y + YHN LHAA V + + + LE A A HDV H
Sbjct 179 YMLTLEDHYHADVAYHNSLHAADVLQSTHVLLATPALDAVFTDLEILAALFAAAIHDVDH 238
Query 114 PGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRILERKSCNIFFLLSVGAFRSVRAKI 173
PG +NQF + LA++YND S LEN H + F++L+ +C+IF LS +S+R +
Sbjct 239 PGVSNQFLINTNSELALMYNDESVLENHHLAVGFKLLQEDNCDIFQNLSKRQRQSLRKMV 298
Query 174 IECI 177
I+ +
Sbjct 299 IDMV 302
> CE28641
Length=626
Score = 75.5 bits (184), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 51/166 (30%), Positives = 80/166 (48%), Gaps = 3/166 (1%)
Query 15 WSFSVLQLDQ-EVNGALFVVGTALLGP--VATDWGCGSNELSQFLQTLQWQYQPNPYHNQ 71
W V ++D+ N +L VV +LL + + + L +L L+ Y+ N YHN
Sbjct 302 WGPDVFKIDELSKNHSLTVVTFSLLRQRNLFKTFEIHQSTLVTYLLNLEHHYRNNHYHNF 361
Query 72 LHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHDVGHPGRTNQFFVACFDPLAII 131
+HAA V + + ++ LE A A HDV HPG TNQ+ + + LAI+
Sbjct 362 IHAADVAQSMHVLLMSPVLTEVFTDLEVLAAIFAGAVHDVDHPGFTNQYLINSNNELAIM 421
Query 132 YNDISPLENFHSCLCFRILERKSCNIFFLLSVGAFRSVRAKIIECI 177
YND S LE H + F++L+ +C+ LS R +I+ +
Sbjct 422 YNDESVLEQHHLAVAFKLLQDSNCDFLANLSRKQRLQFRKIVIDMV 467
> 7297876
Length=605
Score = 74.7 bits (182), Expect = 8e-14, Method: Composition-based stats.
Identities = 54/168 (32%), Positives = 73/168 (43%), Gaps = 5/168 (2%)
Query 15 WSFSVLQLDQEVNGALF-VVGTALLGPVAT--DWGCGSNELSQFLQTLQWQY--QPNPYH 69
W+F V L + +G + V L + + L FL ++ Y NPYH
Sbjct 181 WTFDVFALTEAASGQVVKYVAYELFNRYGSIHKFKIAPGILEAFLHRVEEGYCRYRNPYH 240
Query 70 NQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHDVGHPGRTNQFFVACFDPLA 129
N LHA V G+ + LE A +A L HD H G TN F V A
Sbjct 241 NNLHAVDVMQTIHYCLCNTGLMNWLTDLEIFASLLAALLHDYEHTGTTNNFHVMSGSETA 300
Query 130 IIYNDISPLENFHSCLCFRILERKSCNIFFLLSVGAFRSVRAKIIECI 177
++YND + LEN H+ FR+L NI LS FR +R +IE +
Sbjct 301 LLYNDRAVLENHHASASFRLLREDEYNILSHLSREEFRELRGLVIEMV 348
> CE02038
Length=549
Score = 73.9 bits (180), Expect = 1e-13, Method: Composition-based stats.
Identities = 51/166 (30%), Positives = 80/166 (48%), Gaps = 3/166 (1%)
Query 15 WSFSVLQLDQ-EVNGALFVVGTALLGP--VATDWGCGSNELSQFLQTLQWQYQPNPYHNQ 71
W V ++D+ N +L VV +LL + + + L +L L+ Y+ N YHN
Sbjct 225 WGPDVFKIDELSKNHSLTVVTFSLLRQRNLFKTFEIHQSTLVTYLLNLEHHYRNNHYHNF 284
Query 72 LHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHDVGHPGRTNQFFVACFDPLAII 131
+HAA V + + ++ LE A A HDV HPG TNQ+ + + LAI+
Sbjct 285 IHAADVAQSMHVLLMSPVLTEVFTDLEVLAAIFAGAVHDVDHPGFTNQYLINSNNELAIM 344
Query 132 YNDISPLENFHSCLCFRILERKSCNIFFLLSVGAFRSVRAKIIECI 177
YND S LE H + F++L+ +C+ LS R +I+ +
Sbjct 345 YNDESVLEQHHLAVAFKLLQDSNCDFLANLSRKQRLQFRKIVIDMV 390
> 7291615
Length=1060
Score = 73.9 bits (180), Expect = 2e-13, Method: Composition-based stats.
Identities = 55/174 (31%), Positives = 88/174 (50%), Gaps = 15/174 (8%)
Query 15 WSFSVLQLDQEVN-GALFVVGTALLG--PVATDWGCGSNELSQFLQTLQWQY-QPNPYHN 70
W F + +L++ + L +G + V N +L ++ Y + N YHN
Sbjct 728 WDFDIFKLEEITDYHPLLYLGMEMFRRFDVFATLNIDENVCKAWLAVIEAHYRKSNTYHN 787
Query 71 QLHAA--MVGHGA----VSVANMLGVWQIMEPLEKAALAVATLAHDVGHPGRTNQFFVAC 124
HAA M GA ++ +ML +M+ +E+A +A AHDV HPGR++ F
Sbjct 788 STHAADVMQATGAFITQLTNKDML----VMDRMEEATALIAAAAHDVDHPGRSSAFLCNS 843
Query 125 FDPLAIIYNDISPLENFHSCLCFRI-LERKSCNIFFLLSVGAFRSVRAKIIECI 177
D LA++YND++ LEN H+ + F++ L NIF L ++S R+ II+ I
Sbjct 844 NDALAVLYNDLTVLENHHAAITFKLTLGDDKINIFKNLDKETYKSARSTIIDMI 897
> Hs4826894
Length=634
Score = 71.2 bits (173), Expect = 9e-13, Method: Composition-based stats.
Identities = 56/171 (32%), Positives = 81/171 (47%), Gaps = 8/171 (4%)
Query 14 QWSFSVLQLDQEVNG--ALFVVGTALLG--PVATDWGCGSNELSQFLQTLQWQY--QPNP 67
+WSF V L+ E +G AL + LL + + + + L F++ L+ Y NP
Sbjct 168 KWSFDVFSLN-EASGDHALKFIFYELLTRYDLISRFKIPISALVSFVEALEVGYSKHKNP 226
Query 68 YHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHDVGHPGRTNQFFVACFDP 127
YHN +HAA V + GV + LE A+ + HD H G TN F +
Sbjct 227 YHNLMHAADVTQTVHYLLYKTGVANWLTELEIFAIIFSAAIHDYEHTGTTNNFHIQTRSD 286
Query 128 LAIIYNDISPLENFHSCLCFRILE-RKSCNIFFLLSVGAFRSVRAKIIECI 177
AI+YND S LEN H +R+L+ + NI LS +R R +IE +
Sbjct 287 PAILYNDRSVLENHHLSAAYRLLQDDEEMNILINLSKDDWREFRTLVIEMV 337
> CE24876
Length=562
Score = 66.2 bits (160), Expect = 3e-11, Method: Composition-based stats.
Identities = 44/148 (29%), Positives = 61/148 (41%), Gaps = 25/148 (16%)
Query 55 FLQTLQWQYQPNPYHNQLHAAMVGHGAVSVA-------------------------NMLG 89
F L+ Y PYHN++HAA V HG ++
Sbjct 121 FFHALEKGYWEIPYHNRIHAADVLHGCYYLSAHPVRSTFLTPKTPDSVLTPPHPHHQHSS 180
Query 90 VWQIMEPLEKAALAVATLAHDVGHPGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRI 149
+ + LE AL A HD HPGRTN F V D AI+YND S LEN H+ +++
Sbjct 181 IMSQLSTLELMALFTAAAMHDYDHPGRTNAFLVQVEDKKAILYNDRSVLENHHAAESWKL 240
Query 150 LERKSCNIFFLLSVGAFRSVRAKIIECI 177
L + + L + R ++E I
Sbjct 241 LNKPENHFIENLDPAEMKRFRYLVLEYI 268
> CE01047
Length=918
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 39/136 (28%), Positives = 66/136 (48%), Gaps = 3/136 (2%)
Query 45 WGCGSNELSQFLQTLQWQYQPNPYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAV 104
+ +LS + + Y+P PYHN HA V H + + + +E+ +L +
Sbjct 457 YKLNKRKLSYLVLRVSAGYRPVPYHNWSHAFAVTHFCWLTLRTDAIRRALSDMERLSLLI 516
Query 105 ATLAHDVGHPGRTNQFFVACFD--PLAIIYN-DISPLENFHSCLCFRILERKSCNIFFLL 161
A L HD+ H G TN F + PL+++Y+ + S LE H ++L+++ C+I L
Sbjct 517 ACLCHDIDHRGTTNSFQMQSLQKTPLSVLYSTEGSVLERHHFAQTIKLLQQEECSILENL 576
Query 162 SVGAFRSVRAKIIECI 177
FR++ I E I
Sbjct 577 PAADFRTIVNTIREVI 592
> CE23151
Length=760
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 52/169 (30%), Positives = 78/169 (46%), Gaps = 6/169 (3%)
Query 15 WSFSVLQLDQ-EVNGALFVVGTALLG--PVATDWGCGSNELSQFLQTLQWQYQP-NPYHN 70
W F +L L++ + AL VG + V GC + L +++ +++ Y N YHN
Sbjct 444 WKFDILHLEKVSDHHALSQVGMKVFERWKVCDVLGCSDDLLHRWILSIEAHYHAGNTYHN 503
Query 71 QLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHDVGHPGRTNQFFVACFDPLAI 130
HAA V + V + A +A HD+ HPGR N + + LAI
Sbjct 504 ATHAADVLQATSFFLDSPSVAVHVNESHAVAALLAAAVHDLDHPGRGNAYLINTRQSLAI 563
Query 131 IYNDISPLENFHSCLCFRIL--ERKSCNIFFLLSVGAFRSVRAKIIECI 177
+YND S LEN H L F++ + NIF LS F +R ++E +
Sbjct 564 LYNDNSILENHHIALAFQLTLQHNANVNIFSSLSREEFIQMRHAMVEMV 612
> 7292811
Length=119
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 32/71 (45%), Positives = 41/71 (57%), Gaps = 0/71 (0%)
Query 97 LEKAALAVATLAHDVGHPGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRILERKSCN 156
LE L V+ + HD+ HPG N + + LA+ YNDISPLEN H + FR+LE CN
Sbjct 17 LECLILLVSCICHDLDHPGYNNIYQINARTELALRYNDISPLENHHCSIAFRLLEHPECN 76
Query 157 IFFLLSVGAFR 167
IF S F+
Sbjct 77 IFKNFSRDTFK 87
> Hs4505661
Length=1112
Score = 62.8 bits (151), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 54/214 (25%), Positives = 85/214 (39%), Gaps = 51/214 (23%)
Query 15 WSFSVLQLDQEVNGALFVVGTALLGPVATDWG------CGSNELSQFLQTLQWQYQPNPY 68
W+F + +L +++ + + ++ + D G + + + + L+ Y+ PY
Sbjct 677 WNFPIFELVEKMGEKSGRILSQVMYTLFQDTGLLEIFKIPTQQFMNYFRALENGYRDIPY 736
Query 69 HNQLHAAMVGHGA--VSVANMLGVWQI--------------------------------- 93
HN++HA V H ++ + G+ QI
Sbjct 737 HNRIHATDVLHAVWYLTTRPVPGLQQIHNGCGTGNETDSDGRINHGRIAYISSKSCSNPD 796
Query 94 ---------MEPLEKAALAVATLAHDVGHPGRTNQFFVACFDPLAIIYNDISPLENFHSC 144
+ LE AL VA HD HPGRTN F VA P A++YND S LEN H+
Sbjct 797 ESYGCLSSNIPALELMALYVAAAMHDYDHPGRTNAFLVATNAPQAVLYNDRSVLENHHAA 856
Query 145 LCFRI-LERKSCNIFFLLSVGAFRSVRAKIIECI 177
+ + L R N L F+ R +IE I
Sbjct 857 SAWNLYLSRPEYNFLLHLDHVEFKRFRFLVIEAI 890
> Hs20070127
Length=1141
Score = 61.2 bits (147), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 50/173 (28%), Positives = 67/173 (38%), Gaps = 45/173 (26%)
Query 50 NELSQFLQTLQWQYQPNPYHNQLHAAMVGH-----------GAVSVANMLG--------- 89
E + L+ Y+ PYHN++HA V H G +V N G
Sbjct 733 REFMNYFHALEIGYRDIPYHNRIHATDVLHAVWYLTTQPIPGLSTVINDHGSTSDSDSDS 792
Query 90 -------------VWQIMEP-----------LEKAALAVATLAHDVGHPGRTNQFFVACF 125
+ + + LE AL VA HD HPGRTN F VA
Sbjct 793 GFTHGHMGYVFSKTYNVTDDKYGCLSGNIPALELMALYVAAAMHDYDHPGRTNAFLVATS 852
Query 126 DPLAIIYNDISPLENFHSCLCFRI-LERKSCNIFFLLSVGAFRSVRAKIIECI 177
P A++YND S LEN H+ + + + R N L F+ R +IE I
Sbjct 853 APQAVLYNDRSVLENHHAAAAWNLFMSRPEYNFLINLDHVEFKHFRFLVIEAI 905
> Hs5729972
Length=779
Score = 59.7 bits (143), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 43/155 (27%), Positives = 69/155 (44%), Gaps = 18/155 (11%)
Query 38 LGPVATDW----------GCGSN-----ELSQFLQTLQWQYQPNPYHNQLHAAMVGHGAV 82
+GP W CG++ +L +F+ +++ Y+ PYHN HA V H
Sbjct 469 IGPFENMWPGIFVYMVHRSCGTSCFELEKLCRFIMSVKKNYRRVPYHNWKHAVTVAHCMY 528
Query 83 SVANMLGVWQIMEPLEKAALAVATLAHDVGHPGRTNQFFVACFDPLAIIYNDISPLENFH 142
++ + + LE+ L +A L HD+ H G +N + PLA +Y+ S +E H
Sbjct 529 AI--LQNNHTLFTDLERKGLLIACLCHDLDHRGFSNSYLQKFDHPLAALYS-TSTMEQHH 585
Query 143 SCLCFRILERKSCNIFFLLSVGAFRSVRAKIIECI 177
IL+ + NIF LS + V I + I
Sbjct 586 FSQTVSILQLEGHNIFSTLSSSEYEQVLEIIRKAI 620
> CE04134
Length=393
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 39/129 (30%), Positives = 59/129 (45%), Gaps = 3/129 (2%)
Query 49 SNELSQFLQTLQWQYQPNPYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLA 108
+ +L +F+ T++ Y+ YHN H V H A ++ LE AL V+ L
Sbjct 170 TEDLIRFVLTVRKNYRRVAYHNWAHGWSVAHAMF--ATLMNSPDAFTKLEALALYVSCLC 227
Query 109 HDVGHPGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRILERKSCNIFFLLSVGAFRS 168
HD+ H G+ N + PLA IY+ S +E H IL++ NI LS ++
Sbjct 228 HDLDHRGKNNAYMKTMSTPLASIYS-TSVMERHHFNQTVTILQQDGHNILKSLSSEDYKK 286
Query 169 VRAKIIECI 177
+ I CI
Sbjct 287 TLSLIKHCI 295
> Hs4505657
Length=941
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 36/108 (33%), Positives = 54/108 (50%), Gaps = 1/108 (0%)
Query 52 LSQFLQTLQWQYQPNPYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHDV 111
L++F ++ Y+ PYHN +HA V H + L + +E +E AL ++ + HD+
Sbjct 639 LARFCLMVKKGYRDPPYHNWMHAFSVSHFCYLLYKNLELTNYLEDIEIFALFISCMCHDL 698
Query 112 GHPGRTNQFFVACFDPLAIIY-NDISPLENFHSCLCFRILERKSCNIF 158
H G N F VA LA +Y ++ S +E H IL CNIF
Sbjct 699 DHRGTNNSFQVASKSVLAALYSSEGSVMERHHFAQAIAILNTHGCNIF 746
> Hs20538967
Length=418
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 43/139 (30%), Positives = 61/139 (43%), Gaps = 18/139 (12%)
Query 15 WSFSVLQLDQEVNGALFVVGT----ALLGPVATDWGCGSNELSQFLQTLQWQYQP-NPYH 69
W+F + D+ NG V T +L G + + +L +FL +Q Y NPYH
Sbjct 90 WNFDIFLFDRLTNGNSLVSLTFHLFSLHGLIEY-FHLDMMKLRRFLVMIQEDYHSQNPYH 148
Query 70 NQLHAAMVGHG------AVSVANMLGVWQIMEPLEKAALAVATLAHDVGHPGRTNQFFVA 123
N +HAA V +AN + W I+ L +A HD+ HPG F +
Sbjct 149 NAVHAADVTQAMHCYLKEPKLANSVTPWDILLSL------IAAATHDLDHPGVNQPFLIK 202
Query 124 CFDPLAIIYNDISPLENFH 142
LA +Y + S LEN H
Sbjct 203 TNHYLATLYKNTSVLENHH 221
> Hs9506959
Length=450
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 45/149 (30%), Positives = 64/149 (42%), Gaps = 11/149 (7%)
Query 1 LDEAERNMNAIGKQWSFSVLQLDQEVNGALFVVGTALLGPVATDWGCGSN------ELSQ 54
L +A ++ +G W F + D+ NG V LL + G + L +
Sbjct 102 LGQARHMLSKVG-MWDFDIFLFDRLTNGNSLVT---LLCHLFNTHGLIHHFKLDMVTLHR 157
Query 55 FLQTLQWQYQP-NPYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHDVGH 113
FL +Q Y NPYHN +HAA V + + PL+ +A AHDV H
Sbjct 158 FLVMVQEDYHSQNPYHNAVHAADVTQAMHCYLKEPKLASFLTPLDIMLGLLAAAAHDVDH 217
Query 114 PGRTNQFFVACFDPLAIIYNDISPLENFH 142
PG F + LA +Y ++S LEN H
Sbjct 218 PGVNQPFLIKTNHHLANLYQNMSVLENHH 246
> YOR360c
Length=526
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 46/149 (30%), Positives = 65/149 (43%), Gaps = 21/149 (14%)
Query 11 IGKQWSFSVLQLDQEVNGALFVVGTALLGPVATDWGC--GSNELSQFLQTLQWQY-QPNP 67
I W F L L + L G L+ ++ D N+L L TL+ Y Q N
Sbjct 207 ILSTWDFCALSLSTQ---ELIWCGFTLIKKLSKDAKVLIADNKLLLLLFTLESSYHQVNK 263
Query 68 YHNQLHAAMVGHGAVSVANMLGVWQIM------EPLEKAALAVATLAHDVGHPGRTNQFF 121
+HN HA V M W++ P++ L +A + HDVGHPG NQ
Sbjct 264 FHNFRHAIDV---------MQATWRLCTYLLKDNPVQTLLLCMAAIGHDVGHPGTNNQLL 314
Query 122 VACFDPLAIIYNDISPLENFHSCLCFRIL 150
C +A + ++S LENFH L ++L
Sbjct 315 CNCESEVAQNFKNVSILENFHRELFQQLL 343
> 7298454
Length=1284
Score = 51.2 bits (121), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 36/120 (30%), Positives = 55/120 (45%), Gaps = 2/120 (1%)
Query 52 LSQFLQTLQWQYQPNPYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHDV 111
L ++L +++ Y+ YHN HA V ++ W+I +E AL + L HD+
Sbjct 676 LCRWLLSVKKNYRNVTYHNWRHAFNVAQMMFAILTTTQWWKIFGEIECLALIIGCLCHDL 735
Query 112 GHPGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRILERKSCNIFFLLSVGAF-RSVR 170
H G N F + PLA +Y+ S +E+ H C IL I LS + R +R
Sbjct 736 DHRGTNNSFQIKASSPLAQLYS-TSTMEHHHFDQCLMILNSPGNQILANLSSDDYCRVIR 794
> Hs19923440
Length=934
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 35/115 (30%), Positives = 52/115 (45%), Gaps = 0/115 (0%)
Query 52 LSQFLQTLQWQYQPNPYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHDV 111
L ++L T++ Y+ YHN HA V ++ G I+ +E A+ V L HD+
Sbjct 647 LCRWLLTVRKNYRMVLYHNWRHAFNVCQLMFAMLTTAGFQDILTEVEILAVIVGCLCHDL 706
Query 112 GHPGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRILERKSCNIFFLLSVGAF 166
H G N F LA +Y + LE+ H IL+ + NIF LS +
Sbjct 707 DHRGTNNAFQAKSGSALAQLYGTSATLEHHHFNHAVMILQSEGHNIFANLSSKEY 761
> Hs4585864
Length=860
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 32/107 (29%), Positives = 48/107 (44%), Gaps = 1/107 (0%)
Query 52 LSQFLQTLQWQYQPNPYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHDV 111
L +F+ +L Y+ YHN H VG S+ + + LE A+ A HD+
Sbjct 542 LVRFMYSLSKGYRKITYHNWRHGFNVGQTMFSLLVTGKLKRYFTDLEALAMVTAAFCHDI 601
Query 112 GHPGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRILERKSCNIF 158
H G N + + +PLA ++ S LE H +L +S NIF
Sbjct 602 DHRGTNNLYQMKSQNPLAKLHGS-SILERHHLEFGKTLLRDESLNIF 647
> HsM5453866
Length=858
Score = 45.4 bits (106), Expect = 5e-05, Method: Composition-based stats.
Identities = 33/118 (27%), Positives = 55/118 (46%), Gaps = 1/118 (0%)
Query 52 LSQFLQTLQWQYQPNPYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHDV 111
L++++ T++ Y+ YHN H VG ++ + + LE A+ A HD+
Sbjct 545 LTRWMYTVRKGYRAVTYHNWRHGFNVGQTMFTLLMTGRLKKYYTDLEAFAMLAAAFCHDI 604
Query 112 GHPGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRILERKSCNIFFLLSVGAFRSV 169
H G N + + PLA ++ S LE H +L+ +S NIF L+ F +V
Sbjct 605 DHRGTNNLYQMKSTSPLARLHGS-SILERHHLEYSKTLLQDESLNIFQNLNKRQFETV 661
> Hs21361308
Length=858
Score = 45.4 bits (106), Expect = 5e-05, Method: Composition-based stats.
Identities = 33/118 (27%), Positives = 55/118 (46%), Gaps = 1/118 (0%)
Query 52 LSQFLQTLQWQYQPNPYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHDV 111
L++++ T++ Y+ YHN H VG ++ + + LE A+ A HD+
Sbjct 545 LTRWMYTVRKGYRAVTYHNWQHGFNVGQTMFTLLMTGRLKKYYTDLEAFAMLAAAFCHDI 604
Query 112 GHPGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRILERKSCNIFFLLSVGAFRSV 169
H G N + + PLA ++ S LE H +L+ +S NIF L+ F +V
Sbjct 605 DHRGTNNLYQMKSTSPLARLHGS-SILERHHLEYSKTLLQDESLNIFQNLNKRQFETV 661
> Hs15812214
Length=865
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 34/127 (26%), Positives = 56/127 (44%), Gaps = 2/127 (1%)
Query 52 LSQFLQTLQWQYQPN-PYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHD 110
L +++ +++ Y+ N YHN HA + + + LE AL +A L+HD
Sbjct 585 LCRWILSVKKNYRKNVAYHNWRHAFNTAQCMFAALKAGKIQNKLTDLEILALLIAALSHD 644
Query 111 VGHPGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRILERKSCNIFFLLSVGAFRSVR 170
+ H G N + PLA +Y S +E+ H C IL I LS+ +++
Sbjct 645 LDHRGVNNSYIQRSEHPLAQLYCH-SIMEHHHFDQCLMILNSPGNQILSGLSIEEYKTTL 703
Query 171 AKIIECI 177
I + I
Sbjct 704 KIIKQAI 710
> Hs4505667
Length=875
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 34/127 (26%), Positives = 56/127 (44%), Gaps = 2/127 (1%)
Query 52 LSQFLQTLQWQYQPN-PYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHD 110
L +++ +++ Y+ N YHN HA + + + LE AL +A L+HD
Sbjct 595 LCRWILSVKKNYRKNVAYHNWRHAFNTAQCMFAALKAGKIQNKLTDLEILALLIAALSHD 654
Query 111 VGHPGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRILERKSCNIFFLLSVGAFRSVR 170
+ H G N + PLA +Y S +E+ H C IL I LS+ +++
Sbjct 655 LDHRGVNNSYIQRSEHPLAQLYCH-SIMEHHHFDQCLMILNSPGNQILSGLSIEEYKTTL 713
Query 171 AKIIECI 177
I + I
Sbjct 714 KIIKQAI 720
> Hs15812216
Length=823
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 34/127 (26%), Positives = 56/127 (44%), Gaps = 2/127 (1%)
Query 52 LSQFLQTLQWQYQPN-PYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHD 110
L +++ +++ Y+ N YHN HA + + + LE AL +A L+HD
Sbjct 543 LCRWILSVKKNYRKNVAYHNWRHAFNTAQCMFAALKAGKIQNKLTDLEILALLIAALSHD 602
Query 111 VGHPGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRILERKSCNIFFLLSVGAFRSVR 170
+ H G N + PLA +Y S +E+ H C IL I LS+ +++
Sbjct 603 LDHRGVNNSYIQRSEHPLAQLYCH-SIMEHHHFDQCLMILNSPGNQILSGLSIEEYKTTL 661
Query 171 AKIIECI 177
I + I
Sbjct 662 KIIKQAI 668
> Hs15812212
Length=833
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 33/127 (25%), Positives = 55/127 (43%), Gaps = 2/127 (1%)
Query 52 LSQFLQTLQWQYQPN-PYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHD 110
L +++ +++ Y+ N YHN HA + + + L AL +A L+HD
Sbjct 553 LCRWILSVKKNYRKNVAYHNWRHAFNTAQCMFAALKAGKIQNKLTDLGILALLIAALSHD 612
Query 111 VGHPGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRILERKSCNIFFLLSVGAFRSVR 170
+ H G N + PLA +Y S +E+ H C IL I LS+ +++
Sbjct 613 LDHRGVNNSYIQRSEHPLAQLYCH-SIMEHHHFDQCLMILNSPGNQILSGLSIEEYKTTL 671
Query 171 AKIIECI 177
I + I
Sbjct 672 KIIKQAI 678
> Hs4505669
Length=854
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 29/107 (27%), Positives = 47/107 (43%), Gaps = 1/107 (0%)
Query 52 LSQFLQTLQWQYQPNPYHNQLHAAMVGHGAVSVANMLGVWQIMEPLEKAALAVATLAHDV 111
L +FL ++ Y+ YHN H V ++ + LE A+ A L HD+
Sbjct 540 LVRFLFSISKGYRRITYHNWRHGFNVAQTMFTLLMTGKLKSYYTDLEAFAMVTAGLCHDI 599
Query 112 GHPGRTNQFFVACFDPLAIIYNDISPLENFHSCLCFRILERKSCNIF 158
H G N + + +PLA ++ S LE H +L ++ NI+
Sbjct 600 DHRGTNNLYQMKSQNPLAKLHGS-SILERHHLEFGKFLLSEETLNIY 645
> 7298441
Length=665
Score = 32.7 bits (73), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 16/62 (25%), Positives = 28/62 (45%), Gaps = 0/62 (0%)
Query 24 QEVNGALFVVGTALLGPVATDWGCGSNELSQFLQTLQWQYQPNPYHNQLHAAMVGHGAVS 83
Q + +L+ +G LLG AT+ C + L ++ Y N ++N+ M S
Sbjct 13 QAMTKSLYAIGMGLLGKSATNMDCANTNLESAGKSSSTNYNRNRFYNERFVPMASGTMPS 72
Query 84 VA 85
V+
Sbjct 73 VS 74
> At4g33940_1
Length=399
Score = 32.0 bits (71), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 12/29 (41%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 116 RTNQFFVACFDPLAIIYNDISPLENFHSC 144
R+N+ V CF LA++ DI E F +C
Sbjct 338 RSNRVLVPCFKALAVLIRDIQSTERFRNC 366
> ECU09g0790
Length=350
Score = 31.2 bits (69), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 26/100 (26%), Positives = 39/100 (39%), Gaps = 12/100 (12%)
Query 65 PNPYHNQLHAAMVGHGAVSVANMLGVWQIMEP-LEKAALAVATLAHDVGHPGRTNQFFVA 123
P YHN H + LG P L+K L +A HD+GHPG N+ +
Sbjct 91 PQRYHNLDHLVNTLCFGDFILQRLGEGGYTLPILDKMCLMIALGLHDIGHPGERNRGKIG 150
Query 124 CFD-----PLAIIYNDISPLENFHSCLCFRILERKSCNIF 158
P AI + E H+ +C +++ +F
Sbjct 151 ALSSSYGGPKAISF------ELIHASICRKMVSAYRHTLF 184
> SPBC713.04c
Length=854
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 32/73 (43%), Gaps = 9/73 (12%)
Query 81 AVSVANMLGVWQIMEPLEKAALAVATLAHD-VGHP--------GRTNQFFVACFDPLAII 131
AVS+ ++ VW+ LE+ A L + GH ++FF++ L
Sbjct 112 AVSLGKLIQVWRTPNSLEEREFAPFVLHREYTGHFDDIVSISWSADSRFFISTSKDLTAR 171
Query 132 YNDISPLENFHSC 144
+ + P+E FH C
Sbjct 172 LHSVDPIEGFHPC 184
> Hs19882213
Length=6307
Score = 30.8 bits (68), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 26/43 (60%), Gaps = 2/43 (4%)
Query 79 HGAVSVANMLGVWQIMEPLEKAALAVATLAHDVGHPGRTNQFF 121
+G V+ A M V+++ EPLE+++ A T+ GH GR F+
Sbjct 2349 YGTVAFAQM--VYRVQEPLERSSCANITVRRSGGHFGRLLLFY 2389
> CE28486
Length=1029
Score = 28.9 bits (63), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 26/64 (40%), Gaps = 9/64 (14%)
Query 100 AALAVATLAHDVGHPGRTNQFFVACFDPLAIIYNDISPLE---------NFHSCLCFRIL 150
+ L V + + H GR N C PLA I + S LE +H +C L
Sbjct 303 STLGVDSFVIGMPHRGRLNVLANVCRQPLATILSQFSTLEPADEGSGDVKYHLGVCIERL 362
Query 151 ERKS 154
R+S
Sbjct 363 NRQS 366
Lambda K H
0.325 0.137 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2743263016
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40