bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2704_orf2
Length=130
Score E
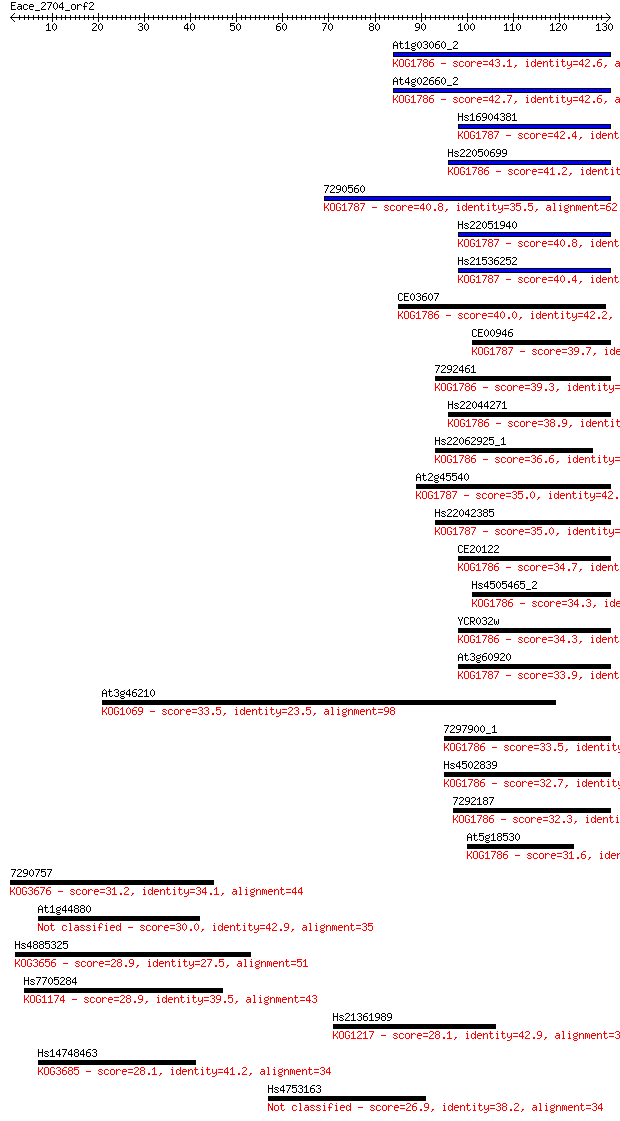
Sequences producing significant alignments: (Bits) Value
At1g03060_2 43.1 1e-04
At4g02660_2 42.7 1e-04
Hs16904381 42.4 2e-04
Hs22050699 41.2 6e-04
7290560 40.8 6e-04
Hs22051940 40.8 6e-04
Hs21536252 40.4 9e-04
CE03607 40.0 0.001
CE00946 39.7 0.002
7292461 39.3 0.002
Hs22044271 38.9 0.003
Hs22062925_1 36.6 0.013
At2g45540 35.0 0.031
Hs22042385 35.0 0.039
CE20122 34.7 0.045
Hs4505465_2 34.3 0.060
YCR032w 34.3 0.068
At3g60920 33.9 0.075
At3g46210 33.5 0.10
7297900_1 33.5 0.11
Hs4502839 32.7 0.17
7292187 32.3 0.25
At5g18530 31.6 0.37
7290757 31.2 0.52
At1g44880 30.0 1.2
Hs4885325 28.9 2.3
Hs7705284 28.9 2.6
Hs21361989 28.1 4.4
Hs14748463 28.1 4.6
Hs4753163 26.9 9.2
> At1g03060_2
Length=717
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 20/47 (42%), Positives = 26/47 (55%), Gaps = 5/47 (10%)
Query 84 RAFMLQASDELLRCMQKRWQEGRLSSFHYLQFLNSAAGRSSSNFSAY 130
R F L A + KRWQ G +S+F YL LN+ AGR S+ + Y
Sbjct 70 RLFKLMA-----KSFSKRWQNGEISNFQYLMHLNTLAGRGYSDLTQY 111
> At4g02660_2
Length=643
Score = 42.7 bits (99), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 20/47 (42%), Positives = 26/47 (55%), Gaps = 5/47 (10%)
Query 84 RAFMLQASDELLRCMQKRWQEGRLSSFHYLQFLNSAAGRSSSNFSAY 130
R F L A + KRWQ G +S+F YL LN+ AGR S+ + Y
Sbjct 70 RLFKLMA-----KSFTKRWQNGEISNFQYLMHLNTLAGRGYSDLTQY 111
> Hs16904381
Length=2863
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 16/33 (48%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 98 MQKRWQEGRLSSFHYLQFLNSAAGRSSSNFSAY 130
M +RWQ +S+F YL FLN+ AGRS ++ + Y
Sbjct 2211 MTQRWQHREISNFEYLMFLNTIAGRSYNDLNQY 2243
> Hs22050699
Length=1606
Score = 41.2 bits (95), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 26/35 (74%), Gaps = 0/35 (0%)
Query 96 RCMQKRWQEGRLSSFHYLQFLNSAAGRSSSNFSAY 130
R M ++WQ+ +S+F YL +LN+AAGR+ +++ Y
Sbjct 1218 RIMLQKWQKRDISNFEYLMYLNTAAGRTCNDYMQY 1252
> 7290560
Length=3614
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 38/66 (57%), Gaps = 5/66 (7%)
Query 69 SLQEAAVGVCCCV--CCRAFMLQASDELLRC--MQKRWQEGRLSSFHYLQFLNSAAGRSS 124
+L VG+ + RA M+ + +L+R M ++WQ +S+F YL FLN+ AGR+
Sbjct 2920 ALPRVGVGIKYGIPQTRRASMM-SPRQLMRNSNMTQKWQRREISNFEYLMFLNTIAGRTY 2978
Query 125 SNFSAY 130
++ + Y
Sbjct 2979 NDLNQY 2984
> Hs22051940
Length=1017
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 98 MQKRWQEGRLSSFHYLQFLNSAAGRSSSNFSAY 130
M +RWQ +S+F YL FLN+ AGR+ ++ + Y
Sbjct 356 MTQRWQRREISNFEYLMFLNTIAGRTYNDLNQY 388
> Hs21536252
Length=2946
Score = 40.4 bits (93), Expect = 9e-04, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 98 MQKRWQEGRLSSFHYLQFLNSAAGRSSSNFSAY 130
M +RWQ +S+F YL FLN+ AGR+ ++ + Y
Sbjct 2285 MTQRWQRREISNFEYLMFLNTIAGRTYNDLNQY 2317
> CE03607
Length=2692
Score = 40.0 bits (92), Expect = 0.001, Method: Composition-based stats.
Identities = 19/45 (42%), Positives = 28/45 (62%), Gaps = 0/45 (0%)
Query 85 AFMLQASDELLRCMQKRWQEGRLSSFHYLQFLNSAAGRSSSNFSA 129
A L A++ L KRW+EG++S+F YL LN AGR+ + S+
Sbjct 2005 ATKLIATEHQLLAYTKRWEEGKMSNFEYLTMLNLFAGRTIHDSSS 2049
> CE00946
Length=2528
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 101 RWQEGRLSSFHYLQFLNSAAGRSSSNFSAY 130
+WQ+ +S+F YL FLN+ AGR+ ++ S Y
Sbjct 1852 KWQKREISNFDYLMFLNTVAGRTFNDLSQY 1881
> 7292461
Length=511
Score = 39.3 bits (90), Expect = 0.002, Method: Composition-based stats.
Identities = 18/40 (45%), Positives = 26/40 (65%), Gaps = 2/40 (5%)
Query 93 ELLRC--MQKRWQEGRLSSFHYLQFLNSAAGRSSSNFSAY 130
ELLR + +RW +S+F YL +LN+ AGRS ++ S Y
Sbjct 187 ELLRASGLTQRWINREISNFEYLMYLNTIAGRSYNDLSQY 226
> Hs22044271
Length=1513
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 23/35 (65%), Gaps = 0/35 (0%)
Query 96 RCMQKRWQEGRLSSFHYLQFLNSAAGRSSSNFSAY 130
+ + +RW+ G +S+F YL LN+ AGRS ++ Y
Sbjct 679 KSVTQRWERGEISNFQYLMHLNTLAGRSYNDLMQY 713
> Hs22062925_1
Length=211
Score = 36.6 bits (83), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 93 ELLRCMQKRWQEGRLSSFHYLQFLNSAAGRSSSN 126
E LR + W GR+S+FHYL LN AGR +
Sbjct 74 EELRSLVLDWVHGRISNFHYLMQLNRLAGRRQGD 107
> At2g45540
Length=2946
Score = 35.0 bits (79), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 27/44 (61%), Gaps = 2/44 (4%)
Query 89 QASDELLRCMQ--KRWQEGRLSSFHYLQFLNSAAGRSSSNFSAY 130
Q ++LLR Q +RW +S+F YL LN+ AGRS ++ + Y
Sbjct 2220 QRPEQLLRRTQLMERWARWEISNFEYLMQLNTLAGRSYNDITQY 2263
> Hs22042385
Length=2040
Score = 35.0 bits (79), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 26/40 (65%), Gaps = 2/40 (5%)
Query 93 ELLRC--MQKRWQEGRLSSFHYLQFLNSAAGRSSSNFSAY 130
E+LR + ++W + +S+F YL LN+ AGR+ ++ S Y
Sbjct 1343 EMLRASGLTQKWVQREISNFEYLMQLNTIAGRTYNDLSQY 1382
> CE20122
Length=931
Score = 34.7 bits (78), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 98 MQKRWQEGRLSSFHYLQFLNSAAGRSSSNFSAY 130
+ +RW G +S+F YL LN+ AGR ++ S Y
Sbjct 162 VTQRWLSGNISNFQYLMHLNTLAGRCYNDLSQY 194
> Hs4505465_2
Length=631
Score = 34.3 bits (77), Expect = 0.060, Method: Composition-based stats.
Identities = 14/30 (46%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 101 RWQEGRLSSFHYLQFLNSAAGRSSSNFSAY 130
+WQ G LS++ YL LN+ A RS ++ S Y
Sbjct 18 QWQRGHLSNYQYLLHLNNLADRSCNDLSQY 47
> YCR032w
Length=2167
Score = 34.3 bits (77), Expect = 0.068, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 22/33 (66%), Gaps = 0/33 (0%)
Query 98 MQKRWQEGRLSSFHYLQFLNSAAGRSSSNFSAY 130
+ K+W G +S+F+YL +N AGRS ++ + Y
Sbjct 1556 ISKKWVRGEISNFYYLLSINILAGRSFNDLTQY 1588
> At3g60920
Length=1857
Score = 33.9 bits (76), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 98 MQKRWQEGRLSSFHYLQFLNSAAGRSSSNFSAY 130
+ +RW +S+F YL LN+ AGRS ++ + Y
Sbjct 1594 LMERWARWEISNFEYLMQLNTLAGRSYNDITQY 1626
> At3g46210
Length=239
Score = 33.5 bits (75), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 23/98 (23%), Positives = 44/98 (44%), Gaps = 20/98 (20%)
Query 21 RPNTATQPLQQETPDNPSQRHRTAICCCCSSSASSSSCCCCCGVWTHVSLQEAAVGVCCC 80
PNT T + Q D+ S + C ++A C V + ++ AV +CCC
Sbjct 97 NPNTTTSVIIQVVHDDGS------LLPCAINAA------CAALVDAGIPMKHLAVAICCC 144
Query 81 VCCRAFMLQASDELLRCMQKRWQEGRLSSFHYLQFLNS 118
+ +++ ++L +E ++++F YL F N+
Sbjct 145 LAENGYLVLDPNKL--------EEKKMTAFAYLVFPNT 174
> 7297900_1
Length=917
Score = 33.5 bits (75), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 18/36 (50%), Positives = 24/36 (66%), Gaps = 1/36 (2%)
Query 95 LRCMQKRWQEGRLSSFHYLQFLNSAAGRSSSNFSAY 130
LR + W G+LS+F YL LN+A GRS +N +AY
Sbjct 341 LREYTEMWCNGQLSNFDYLTILNNACGRSLTN-AAY 375
> Hs4502839
Length=3801
Score = 32.7 bits (73), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 12/36 (33%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 95 LRCMQKRWQEGRLSSFHYLQFLNSAAGRSSSNFSAY 130
+ + W G++++F YL LN AGRS ++ Y
Sbjct 3128 ITALTNLWYTGQITNFEYLTHLNKHAGRSFNDLMQY 3163
> 7292187
Length=2517
Score = 32.3 bits (72), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 11/34 (32%), Positives = 22/34 (64%), Gaps = 0/34 (0%)
Query 97 CMQKRWQEGRLSSFHYLQFLNSAAGRSSSNFSAY 130
+ ++W+EG L+++ YL LN +GR+ ++ Y
Sbjct 1776 AITQQWREGLLTNWEYLMTLNQISGRTYNDLMQY 1809
> At5g18530
Length=1598
Score = 31.6 bits (70), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 12/23 (52%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 100 KRWQEGRLSSFHYLQFLNSAAGR 122
+W +G LS+F YL LN AGR
Sbjct 349 DKWWKGELSNFEYLLVLNKLAGR 371
> 7290757
Length=1123
Score = 31.2 bits (69), Expect = 0.52, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 23/44 (52%), Gaps = 5/44 (11%)
Query 1 PLQQQSKRPSKASKQHRRPSRPNTATQPLQQETPDNPSQRHRTA 44
P Q ++ R +K+++ RR P Q + PD SQR R+A
Sbjct 1055 PAQDEAARRAKSARVRRR-----NKVSPEQSDDPDERSQRGRSA 1093
> At1g44880
Length=1038
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 18/39 (46%), Gaps = 4/39 (10%)
Query 7 KRPSKASKQ----HRRPSRPNTATQPLQQETPDNPSQRH 41
KR +K S RP R T P+Q +TP SQ H
Sbjct 457 KRSTKLSSSFVACSSRPKRKKTMEVPIQSQTPQTSSQSH 495
> Hs4885325
Length=330
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 14/51 (27%), Positives = 22/51 (43%), Gaps = 0/51 (0%)
Query 2 LQQQSKRPSKASKQHRRPSRPNTATQPLQQETPDNPSQRHRTAICCCCSSS 52
L + P + P+ N+ P+ + Q+ A+CCCCSSS
Sbjct 268 LLGDAHSPPLYTYLTLLPATYNSMINPIIYAFRNQDVQKVLWAVCCCCSSS 318
> Hs7705284
Length=565
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 17/43 (39%), Positives = 22/43 (51%), Gaps = 2/43 (4%)
Query 4 QQSKRPSKASKQHRRPSRPNTATQPLQQETPDNPSQRHRTAIC 46
QQ K SK SK RPS N+A+ P Q P +++ A C
Sbjct 71 QQKKALSKTSKV--RPSTGNSASTPQSQCLPSEIEVKYKMAEC 111
> Hs21361989
Length=1316
Score = 28.1 bits (61), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 18/37 (48%), Gaps = 2/37 (5%)
Query 71 QEAAVGVCCCVCCR-AFMLQASDELLRCMQ-KRWQEG 105
Q A G C V CR F+L E+LRC +W G
Sbjct 284 QPAKFGTICYVSCRQGFILSGVKEMLRCTTSGKWNVG 320
> Hs14748463
Length=1455
Score = 28.1 bits (61), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 23/39 (58%), Gaps = 5/39 (12%)
Query 7 KRPSKASKQHRRPSRP-----NTATQPLQQETPDNPSQR 40
K P++A+ +RP+ P N+AT+P E P+NP+
Sbjct 350 KPPTQAAYIAQRPNDPGRSRQNSATRPDNSEIPENPAME 388
> Hs4753163
Length=3144
Score = 26.9 bits (58), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 18/38 (47%), Gaps = 4/38 (10%)
Query 57 SCCCCC----GVWTHVSLQEAAVGVCCCVCCRAFMLQA 90
CCC G+W+ VS E C + C F+L+A
Sbjct 2282 DCCCLALQLPGLWSVVSSTEFVTHACSLIYCVHFILEA 2319
Lambda K H
0.322 0.126 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1246445644
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40