bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2692_orf1
Length=54
Score E
Sequences producing significant alignments: (Bits) Value
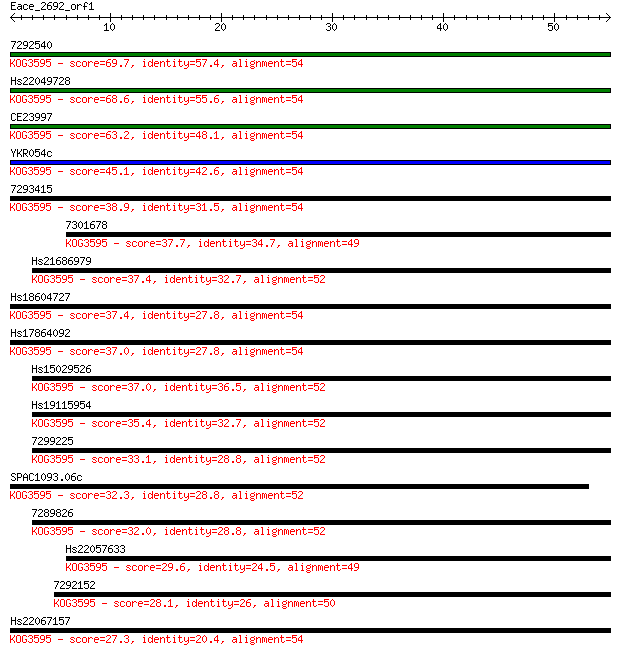
7292540 69.7 1e-12
Hs22049728 68.6 3e-12
CE23997 63.2 1e-10
YKR054c 45.1 4e-05
7293415 38.9 0.002
7301678 37.7 0.005
Hs21686979 37.4 0.006
Hs18604727 37.4 0.007
Hs17864092 37.0 0.008
Hs15029526 37.0 0.009
Hs19115954 35.4 0.023
7299225 33.1 0.13
SPAC1093.06c 32.3 0.25
7289826 32.0 0.26
Hs22057633 29.6 1.6
7292152 28.1 4.1
Hs22067157 27.3 6.3
> 7292540
Length=4680
Score = 69.7 bits (169), Expect = 1e-12, Method: Composition-based stats.
Identities = 31/54 (57%), Positives = 40/54 (74%), Gaps = 0/54 (0%)
Query 1 MNGLSIFQIKAGRNYNTASFEQDLRVVMKRAALKEEKIAFVIDESNALGPAFLE 54
MNGLSIFQIK Y + F++DLR V++R+ K+EKIAF++DESN L FLE
Sbjct 2922 MNGLSIFQIKVHNKYTSEDFDEDLRCVLRRSGCKDEKIAFILDESNVLDSGFLE 2975
> Hs22049728
Length=2274
Score = 68.6 bits (166), Expect = 3e-12, Method: Composition-based stats.
Identities = 30/54 (55%), Positives = 39/54 (72%), Gaps = 0/54 (0%)
Query 1 MNGLSIFQIKAGRNYNTASFEQDLRVVMKRAALKEEKIAFVIDESNALGPAFLE 54
MNGLS++QIK R Y F++DLR V++R+ K EKIAF++DESN L FLE
Sbjct 581 MNGLSVYQIKVHRKYTGEDFDEDLRTVLRRSGCKNEKIAFIMDESNVLDSGFLE 634
> CE23997
Length=4568
Score = 63.2 bits (152), Expect = 1e-10, Method: Composition-based stats.
Identities = 26/54 (48%), Positives = 38/54 (70%), Gaps = 0/54 (0%)
Query 1 MNGLSIFQIKAGRNYNTASFEQDLRVVMKRAALKEEKIAFVIDESNALGPAFLE 54
+NGLS+FQ+K Y A F++D+R V++RA + EK+ F++DESN L FLE
Sbjct 2896 LNGLSVFQLKVHSKYTAADFDEDMRTVLRRAGCRNEKLCFIMDESNMLDTGFLE 2949
> YKR054c
Length=4092
Score = 45.1 bits (105), Expect = 4e-05, Method: Composition-based stats.
Identities = 23/54 (42%), Positives = 32/54 (59%), Gaps = 0/54 (0%)
Query 1 MNGLSIFQIKAGRNYNTASFEQDLRVVMKRAALKEEKIAFVIDESNALGPAFLE 54
+NGL I Q K R+ N + F+ L+ + +LKE + +IDESN L AFLE
Sbjct 2776 LNGLKIVQPKIHRHSNLSDFDMILKKAISDCSLKESRTCLIIDESNILETAFLE 2829
> 7293415
Length=4081
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 33/54 (61%), Gaps = 0/54 (0%)
Query 1 MNGLSIFQIKAGRNYNTASFEQDLRVVMKRAALKEEKIAFVIDESNALGPAFLE 54
+N + +QI+ RNY+ +F +DLRV+ + A + + + F++ +S + FLE
Sbjct 2398 VNEYNCWQIEMRRNYDLNAFHEDLRVLYRIAGIDNQPVTFLLIDSQIVEEEFLE 2451
> 7301678
Length=4820
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 28/49 (57%), Gaps = 0/49 (0%)
Query 6 IFQIKAGRNYNTASFEQDLRVVMKRAALKEEKIAFVIDESNALGPAFLE 54
+F+I R YN A+F +DL+V+ A +K +K+ F+ + FLE
Sbjct 3388 VFEITISRGYNEAAFREDLKVLYTIAGVKRKKVVFLFTGAQVAEEGFLE 3436
> Hs21686979
Length=551
Score = 37.4 bits (85), Expect = 0.006, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 29/52 (55%), Gaps = 0/52 (0%)
Query 3 GLSIFQIKAGRNYNTASFEQDLRVVMKRAALKEEKIAFVIDESNALGPAFLE 54
G QI+ R YN SF +DLR + K A ++++ + F+ ++ + FLE
Sbjct 437 GYKCLQIELSRGYNYDSFHEDLRKLYKMAGVEDKNMVFLFTDTQIVVEEFLE 488
> Hs18604727
Length=2504
Score = 37.4 bits (85), Expect = 0.007, Method: Composition-based stats.
Identities = 15/54 (27%), Positives = 31/54 (57%), Gaps = 0/54 (0%)
Query 1 MNGLSIFQIKAGRNYNTASFEQDLRVVMKRAALKEEKIAFVIDESNALGPAFLE 54
M+G +F K R Y F+ DL+ V++ A ++ +++ ++++ + P FLE
Sbjct 2441 MHGAVLFSPKISRGYELKQFKNDLKHVLQLAGIEAQQVVLLLEDYQFVHPTFLE 2494
> Hs17864092
Length=4024
Score = 37.0 bits (84), Expect = 0.008, Method: Composition-based stats.
Identities = 15/54 (27%), Positives = 32/54 (59%), Gaps = 0/54 (0%)
Query 1 MNGLSIFQIKAGRNYNTASFEQDLRVVMKRAALKEEKIAFVIDESNALGPAFLE 54
M S+FQ++ + Y+T + +DL+V++++ A E + F+ ++ +FLE
Sbjct 2357 MADYSVFQVEISKGYDTTEWHEDLKVILRKCAEGEMQGVFLFTDTQIKEESFLE 2410
> Hs15029526
Length=4490
Score = 37.0 bits (84), Expect = 0.009, Method: Composition-based stats.
Identities = 19/52 (36%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 3 GLSIFQIKAGRNYNTASFEQDLRVVMKRAALKEEKIAFVIDESNALGPAFLE 54
G IFQI R+YN + DL+ + K A + I F+ +S AFLE
Sbjct 2838 GYQIFQITLTRSYNVTNLTDDLKALYKVAGADGKGITFIFTDSEIKDEAFLE 2889
> Hs19115954
Length=4624
Score = 35.4 bits (80), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 30/52 (57%), Gaps = 0/52 (0%)
Query 3 GLSIFQIKAGRNYNTASFEQDLRVVMKRAALKEEKIAFVIDESNALGPAFLE 54
G FQI R+YNT++ +DL+V+ + A + + I F+ ++ +FLE
Sbjct 2974 GYVSFQITLTRSYNTSNLMEDLKVLYRTAGQQGKGITFIFTDNEIKDESFLE 3025
> 7299225
Length=3508
Score = 33.1 bits (74), Expect = 0.13, Method: Composition-based stats.
Identities = 15/52 (28%), Positives = 28/52 (53%), Gaps = 0/52 (0%)
Query 3 GLSIFQIKAGRNYNTASFEQDLRVVMKRAALKEEKIAFVIDESNALGPAFLE 54
G FQI R+YN A+F +DL+++ + ++ + F+ + + FLE
Sbjct 2961 GYKTFQIALTRSYNVANFLEDLKLLYRTCGVQGKGTTFLFTDMDIKEEGFLE 3012
> SPAC1093.06c
Length=1889
Score = 32.3 bits (72), Expect = 0.25, Method: Composition-based stats.
Identities = 15/52 (28%), Positives = 27/52 (51%), Gaps = 0/52 (0%)
Query 1 MNGLSIFQIKAGRNYNTASFEQDLRVVMKRAALKEEKIAFVIDESNALGPAF 52
+N S+F+++ + Y+ FE +L+ ++ A K I+ES A P F
Sbjct 569 LNSFSLFELQKNQTYSIEDFEDNLKSILILAGTTNCKACLAINESIAGVPGF 620
> 7289826
Length=507
Score = 32.0 bits (71), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 15/52 (28%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 3 GLSIFQIKAGRNYNTASFEQDLRVVMKRAALKEEKIAFVIDESNALGPAFLE 54
G FQ+ R+YNT + +DL+ + + A L + F+ + +FLE
Sbjct 152 GYKFFQMTLTRSYNTGNLTEDLKFLYRTAGLDGNGMTFIFTANEIKEESFLE 203
> Hs22057633
Length=1770
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 12/49 (24%), Positives = 27/49 (55%), Gaps = 0/49 (0%)
Query 6 IFQIKAGRNYNTASFEQDLRVVMKRAALKEEKIAFVIDESNALGPAFLE 54
+F+I R Y+ SF +DL+ + + ++ + + F+ +++ FLE
Sbjct 1281 VFEILLSRGYSENSFREDLKSLYLKLGIENKAMIFLFTDAHVAEEGFLE 1329
> 7292152
Length=3868
Score = 28.1 bits (61), Expect = 4.1, Method: Composition-based stats.
Identities = 13/50 (26%), Positives = 24/50 (48%), Gaps = 0/50 (0%)
Query 5 SIFQIKAGRNYNTASFEQDLRVVMKRAALKEEKIAFVIDESNALGPAFLE 54
S FQ + +NY + D++ ++K A + F+I E+ FL+
Sbjct 2301 SFFQPEITKNYGANDWHDDIKAILKEAGGMNKHTTFLITENQIKMELFLQ 2350
> Hs22067157
Length=3672
Score = 27.3 bits (59), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 11/54 (20%), Positives = 28/54 (51%), Gaps = 0/54 (0%)
Query 1 MNGLSIFQIKAGRNYNTASFEQDLRVVMKRAALKEEKIAFVIDESNALGPAFLE 54
MN ++QI+ +NY + +DL+ ++ + + + F+ ++ +F+E
Sbjct 2006 MNAYELYQIEITKNYAGNDWREDLKKIILQVGVATKSTVFLFADNQIKDESFVE 2059
Lambda K H
0.319 0.134 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1200194442
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40