bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2588_orf1
Length=115
Score E
Sequences producing significant alignments: (Bits) Value
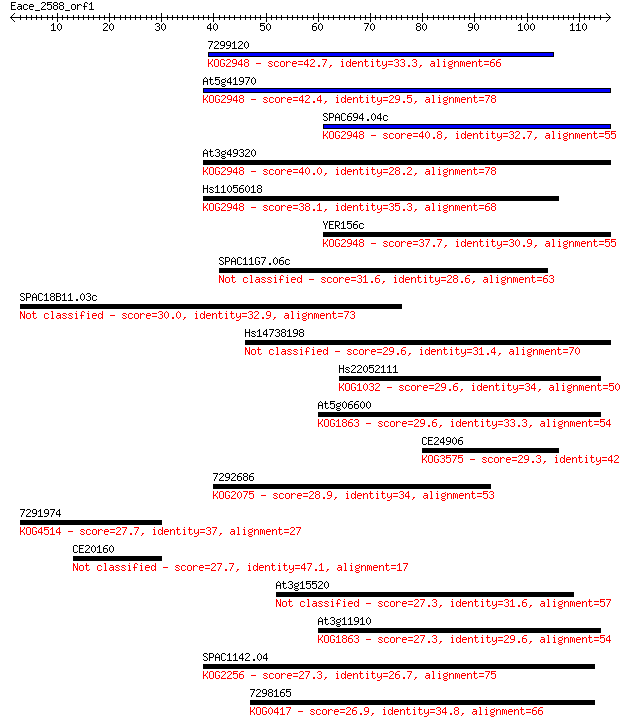
7299120 42.7 2e-04
At5g41970 42.4 2e-04
SPAC694.04c 40.8 6e-04
At3g49320 40.0 0.001
Hs11056018 38.1 0.004
YER156c 37.7 0.005
SPAC11G7.06c 31.6 0.40
SPAC18B11.03c 30.0 1.1
Hs14738198 29.6 1.2
Hs22052111 29.6 1.4
At5g06600 29.6 1.5
CE24906 29.3 2.0
7292686 28.9 2.3
7291974 27.7 5.5
CE20160 27.7 5.7
At3g15520 27.3 6.3
At3g11910 27.3 6.5
SPAC1142.04 27.3 7.7
7298165 26.9 9.6
> 7299120
Length=315
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 39/66 (59%), Gaps = 1/66 (1%)
Query 39 FRLALAYADETFRDAVIYLRDKWLPARNIVKQAILNSPAPKEFQEHVLELAKWAPYEDHV 98
FR A+ A F D V+ + W+PAR+ V++A+ N+ E +L L + P++ H+
Sbjct 162 FRQAMDTAGREFVDNVVEVSCSWIPARDHVREALKNAKNVHPTGE-ILVLKNFCPWKSHL 220
Query 99 YNLERE 104
++LE+E
Sbjct 221 FDLEKE 226
> At5g41970
Length=346
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 45/78 (57%), Gaps = 1/78 (1%)
Query 38 AFRLALAYADETFRDAVIYLRDKWLPARNIVKQAILNSPAPKEFQEHVLELAKWAPYEDH 97
AF+ A+A A + F ++V + WLPAR+IV Q L + ++ L ++ P++ H
Sbjct 194 AFQRAMALAGKEFLESVQFHARSWLPARSIVMQC-LEERFKTDPSGEIMILDRFCPWKLH 252
Query 98 VYNLERELGLSEKLKFAV 115
++ LE+E+ + +K+ +
Sbjct 253 LFELEQEMKIEPLIKYVI 270
> SPAC694.04c
Length=324
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 38/58 (65%), Gaps = 11/58 (18%)
Query 61 WLPARNIVKQAIL---NSPAPKEFQEHVLELAKWAPYEDHVYNLERELGLSEKLKFAV 115
WLPA+ + ++AIL +SP +L + ++ P++ H++++E+ELG+ + K+A+
Sbjct 198 WLPAKTLAREAILKAKDSP--------ILIVDQFFPWKGHLFDIEKELGIENQFKYAI 247
> At3g49320
Length=354
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/78 (28%), Positives = 41/78 (52%), Gaps = 1/78 (1%)
Query 38 AFRLALAYADETFRDAVIYLRDKWLPARNIVKQAILNSPAPKEFQEHVLELAKWAPYEDH 97
AF A+ A F + V + WLPAR+IV + + E +++L+K P++ H
Sbjct 202 AFHRAMELAGSEFLECVHFHAKSWLPARSIVMECLAKRYDIDSSGE-IMKLSKQCPWKLH 260
Query 98 VYNLERELGLSEKLKFAV 115
++ LE E+ + +K+ +
Sbjct 261 IFELEEEMKIDPPIKYVL 278
> Hs11056018
Length=376
Score = 38.1 bits (87), Expect = 0.004, Method: Composition-based stats.
Identities = 24/73 (32%), Positives = 41/73 (56%), Gaps = 10/73 (13%)
Query 38 AFRLALAYADETFRDAVIYLRDKWLPARNIVKQAILNSPAPKEFQ----EHVLELAKWA- 92
F+ A+ E F + + + WLPAR +V++A+ + FQ ++ELAK A
Sbjct 216 GFKRAMDLVQEEFLQRLDFYQHSWLPARALVEEAL-----AQRFQVDPSGEIVELAKGAC 270
Query 93 PYEDHVYNLEREL 105
P+++H+Y+LE L
Sbjct 271 PWKEHLYHLESGL 283
> YER156c
Length=338
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 35/55 (63%), Gaps = 1/55 (1%)
Query 61 WLPARNIVKQAILNSPAPKEFQEHVLELAKWAPYEDHVYNLERELGLSEKLKFAV 115
WLPA+ +V QAI + + ++ L ++ P+++H+Y LERE + ++++F +
Sbjct 211 WLPAKALVAQAI-DERMDVDKSGKIIVLPQFCPWKEHLYELEREKNIEKQIEFVL 264
> SPAC11G7.06c
Length=430
Score = 31.6 bits (70), Expect = 0.40, Method: Composition-based stats.
Identities = 18/63 (28%), Positives = 32/63 (50%), Gaps = 5/63 (7%)
Query 41 LALAYADETFRDAVIYLRDKWLPARNIVKQAILNSPAPKEFQEHVLELAKWAPYEDHVYN 100
LA+ Y+ +T+ +LR+ P + +V +L + P++FQ+ A W + HV N
Sbjct 151 LAINYSPKTYTKIFDFLRNGRAPLQVLVHHCMLYNFYPEDFQD-----ALWCAVQAHVMN 205
Query 101 LER 103
R
Sbjct 206 QVR 208
> SPAC18B11.03c
Length=440
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 24/79 (30%), Positives = 40/79 (50%), Gaps = 10/79 (12%)
Query 3 SAPASSSAAAKGKGWAPACSFVDIPIIGG---SAEALCAFRLALAYAD---ETFRDAVIY 56
S PA SS K +A CS + P + A+ C+F L++ + D E + I
Sbjct 366 SFPADSSVKIKNMAFAQPCSSLSAPFVLNVITVADGPCSFSLSI-FDDGNTEQTHELAIK 424
Query 57 LRDKWLPARNIVKQAILNS 75
+RDK+L +I+++ +NS
Sbjct 425 IRDKFL---SILEKVSMNS 440
> Hs14738198
Length=1214
Score = 29.6 bits (65), Expect = 1.2, Method: Composition-based stats.
Identities = 22/70 (31%), Positives = 36/70 (51%), Gaps = 13/70 (18%)
Query 46 ADETFRDAVIYLRDKWLPARNIVKQAILNSPAPKEFQEHVLELAKWAPYEDHVYNLEREL 105
ADET D+ A N+ + +++PAP++FQ ++ LA +ED N RE
Sbjct 804 ADET--------EDERSEAENVAENFSISNPAPQQFQ-GIINLA----FEDATENECREF 850
Query 106 GLSEKLKFAV 115
++K K +V
Sbjct 851 SATKKFKRSV 860
> Hs22052111
Length=650
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 25/50 (50%), Gaps = 0/50 (0%)
Query 64 ARNIVKQAILNSPAPKEFQEHVLELAKWAPYEDHVYNLERELGLSEKLKF 113
AR V I P + ++E W+ ED+ ++LEREL +EKL
Sbjct 415 ARLRVSSEIRYRKQPWSLVKSLIEKNSWSGIEDYFHHLERELAKAEKLSL 464
> At5g06600
Length=1115
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 18/54 (33%), Positives = 28/54 (51%), Gaps = 3/54 (5%)
Query 60 KWLPARNIVKQAILNSPAPKEFQEHVLELAKWAPYEDHVYNLERELGLSEKLKF 113
+++ R +V+ L P EF VLEL+K Y+D V + +LGL + K
Sbjct 795 EYVQNRQLVRFRALEKPKEDEF---VLELSKQHTYDDVVEKVAEKLGLDDPSKL 845
> CE24906
Length=435
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 11/26 (42%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 80 EFQEHVLELAKWAPYEDHVYNLEREL 105
+FQ ++ AKW Y D + NL+ EL
Sbjct 224 DFQTYMCAAAKWLSYSDRIRNLDDEL 249
> 7292686
Length=291
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 30/60 (50%), Gaps = 7/60 (11%)
Query 40 RLALAYADETFRDAVIYLRDKWLPARNI-------VKQAILNSPAPKEFQEHVLELAKWA 92
++A YA++ D V+ + K PA + V Q +L +P E +HV+EL + A
Sbjct 132 KIANLYAEQLMSDIVLLVDGKEFPAHRVILCASSDVFQVMLMNPEWNECSKHVIELHEEA 191
> 7291974
Length=302
Score = 27.7 bits (60), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 10/27 (37%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 3 SAPASSSAAAKGKGWAPACSFVDIPII 29
S P+ +A + G G PA S+ +IP +
Sbjct 21 SGPSEGAARSSGSGATPASSYTEIPFL 47
> CE20160
Length=325
Score = 27.7 bits (60), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 8/17 (47%), Positives = 14/17 (82%), Gaps = 0/17 (0%)
Query 13 KGKGWAPACSFVDIPII 29
K GW+P+C+F+DI ++
Sbjct 122 KAAGWSPSCAFLDIVLL 138
> At3g15520
Length=461
Score = 27.3 bits (59), Expect = 6.3, Method: Composition-based stats.
Identities = 18/60 (30%), Positives = 32/60 (53%), Gaps = 3/60 (5%)
Query 52 DAVIYLRDKWLPARN--IVKQAILNSPAPKEFQ-EHVLELAKWAPYEDHVYNLERELGLS 108
+AV+Y D +P +++AI +P+ K Q H L L+ W PY N+++ L ++
Sbjct 113 EAVLYSPDTKVPRTGELALRRAIPANPSMKIIQARHSLRLSMWKPYGTMESNVKKALKVA 172
> At3g11910
Length=1124
Score = 27.3 bits (59), Expect = 6.5, Method: Composition-based stats.
Identities = 16/54 (29%), Positives = 27/54 (50%), Gaps = 3/54 (5%)
Query 60 KWLPARNIVKQAILNSPAPKEFQEHVLELAKWAPYEDHVYNLERELGLSEKLKF 113
+++ R +V+ L P EF +EL+K Y+D V + +LGL + K
Sbjct 804 EYVQNRELVRFRTLEKPKEDEF---TMELSKLHTYDDVVERVAEKLGLDDPSKL 854
> SPAC1142.04
Length=707
Score = 27.3 bits (59), Expect = 7.7, Method: Composition-based stats.
Identities = 20/75 (26%), Positives = 32/75 (42%), Gaps = 0/75 (0%)
Query 38 AFRLALAYADETFRDAVIYLRDKWLPARNIVKQAILNSPAPKEFQEHVLELAKWAPYEDH 97
A L L A+ + Y+R + RN + Q +S P + +V L WA
Sbjct 387 AVNLFLLDAESCYLIGFRYIRQLAITLRNTIHQPSKDSRKPVQSWSYVHSLDFWARLLSQ 446
Query 98 VYNLERELGLSEKLK 112
L RE G++ +L+
Sbjct 447 AAWLSREKGVASELQ 461
> 7298165
Length=151
Score = 26.9 bits (58), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 31/72 (43%), Gaps = 8/72 (11%)
Query 47 DETFRDAVIYLRDKWLPARNIVK-----QAILNSPAPKE-FQEHVLELAKWAPYEDHVYN 100
D R + L+DKW PA I QA+L++P P + V EL W E
Sbjct 81 DRVGRICLDILKDKWSPALQIRTVLLSIQALLSAPNPDDPLANDVAEL--WKVNERRAIQ 138
Query 101 LERELGLSEKLK 112
L RE L ++
Sbjct 139 LARECTLKHAMQ 150
Lambda K H
0.318 0.132 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1178471386
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40