bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2582_orf3
Length=122
Score E
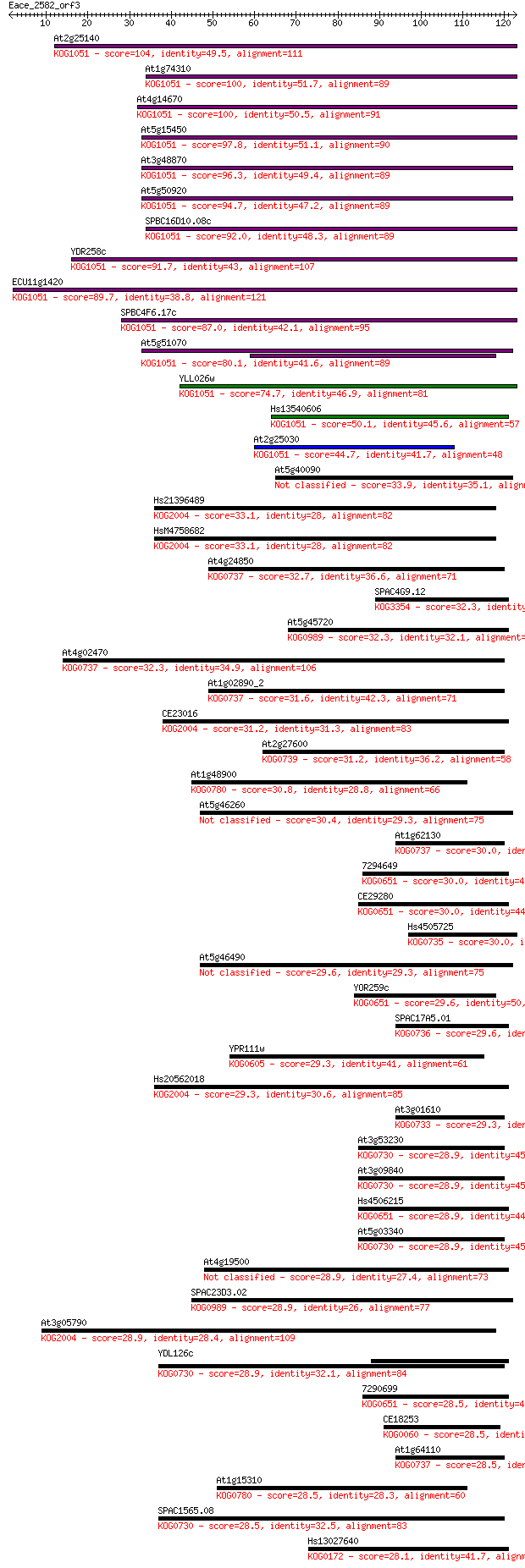
Sequences producing significant alignments: (Bits) Value
At2g25140 104 4e-23
At1g74310 100 5e-22
At4g14670 100 8e-22
At5g15450 97.8 4e-21
At3g48870 96.3 1e-20
At5g50920 94.7 4e-20
SPBC16D10.08c 92.0 2e-19
YDR258c 91.7 3e-19
ECU11g1420 89.7 1e-18
SPBC4F6.17c 87.0 8e-18
At5g51070 80.1 1e-15
YLL026w 74.7 4e-14
Hs13540606 50.1 1e-06
At2g25030 44.7 5e-05
At5g40090 33.9 0.075
Hs21396489 33.1 0.13
HsM4758682 33.1 0.14
At4g24850 32.7 0.15
SPAC4G9.12 32.3 0.20
At5g45720 32.3 0.20
At4g02470 32.3 0.24
At1g02890_2 31.6 0.37
CE23016 31.2 0.43
At2g27600 31.2 0.55
At1g48900 30.8 0.59
At5g46260 30.4 0.81
At1g62130 30.0 1.0
7294649 30.0 1.1
CE29280 30.0 1.1
Hs4505725 30.0 1.2
At5g46490 29.6 1.3
YOR259c 29.6 1.4
SPAC17A5.01 29.6 1.6
YPR111w 29.3 1.7
Hs20562018 29.3 1.8
At3g01610 29.3 2.0
At3g53230 28.9 2.3
At3g09840 28.9 2.3
Hs4506215 28.9 2.5
At5g03340 28.9 2.5
At4g19500 28.9 2.6
SPAC23D3.02 28.9 2.6
At3g05790 28.9 2.7
YDL126c 28.9 2.8
7290699 28.5 3.0
CE18253 28.5 3.1
At1g64110 28.5 3.1
At1g15310 28.5 3.2
SPAC1565.08 28.5 3.5
Hs13027640 28.1 3.7
> At2g25140
Length=874
Score = 104 bits (260), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 55/118 (46%), Positives = 76/118 (64%), Gaps = 7/118 (5%)
Query 12 RQLAALEELTANGRRLALSLH-------DVAHILHLWTGIPLGKMTEDEISRVLRLADIL 64
RQL E+ N R+ SL D+A I+ WTGIPL + + E +++ L ++L
Sbjct 502 RQLEEAEKNLTNFRQFGQSLLREVVTDLDIAEIVSKWTGIPLSNLQQSEREKLVMLEEVL 561
Query 65 SSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFD 122
RVIGQ AVK+VADA+ RAGLS N+P+ +FMF+G +GVGKTELAKA+A +F+
Sbjct 562 HHRVIGQDMAVKSVADAIRRSRAGLSDPNRPIASFMFMGPTGVGKTELAKALAGYLFN 619
> At1g74310
Length=911
Score = 100 bits (250), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 46/89 (51%), Positives = 67/89 (75%), Gaps = 0/89 (0%)
Query 34 VAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPKN 93
+A ++ WTGIP+ ++ ++E R++ LAD L RV+GQ QAV AV++A+ RAGL
Sbjct 537 IAEVVSRWTGIPVTRLGQNEKERLIGLADRLHKRVVGQNQAVNAVSEAILRSRAGLGRPQ 596
Query 94 KPLGTFMFLGSSGVGKTELAKAVAEEMFD 122
+P G+F+FLG +GVGKTELAKA+AE++FD
Sbjct 597 QPTGSFLFLGPTGVGKTELAKALAEQLFD 625
> At4g14670
Length=623
Score = 100 bits (248), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 46/91 (50%), Positives = 67/91 (73%), Gaps = 0/91 (0%)
Query 32 HDVAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSP 91
++A ++ WTGIP+ ++ ++E R++ LAD L RV+GQ +AVKAVA A+ R GL
Sbjct 500 ENIAEVVSRWTGIPVTRLDQNEKKRLISLADKLHERVVGQDEAVKAVAAAILRSRVGLGR 559
Query 92 KNKPLGTFMFLGSSGVGKTELAKAVAEEMFD 122
+P G+F+FLG +GVGKTELAKA+AE++FD
Sbjct 560 PQQPSGSFLFLGPTGVGKTELAKALAEQLFD 590
> At5g15450
Length=968
Score = 97.8 bits (242), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 46/90 (51%), Positives = 64/90 (71%), Gaps = 0/90 (0%)
Query 33 DVAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPK 92
D+A I+ WTGIP+ K+ + E ++L L + L RV+GQ AV AVA+A+ RAGLS
Sbjct 615 DIAEIVSKWTGIPVSKLQQSERDKLLHLEEELHKRVVGQNPAVTAVAEAIQRSRAGLSDP 674
Query 93 NKPLGTFMFLGSSGVGKTELAKAVAEEMFD 122
+P+ +FMF+G +GVGKTELAKA+A MF+
Sbjct 675 GRPIASFMFMGPTGVGKTELAKALASYMFN 704
> At3g48870
Length=952
Score = 96.3 bits (238), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 44/89 (49%), Positives = 64/89 (71%), Gaps = 0/89 (0%)
Query 33 DVAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPK 92
D+ HI+ WTGIP+ K++ DE SR+L++ L +RVIGQ +AVKA++ A+ R GL
Sbjct 596 DIQHIVATWTGIPVEKVSSDESSRLLQMEQTLHTRVIGQDEAVKAISRAIRRARVGLKNP 655
Query 93 NKPLGTFMFLGSSGVGKTELAKAVAEEMF 121
N+P+ +F+F G +GVGK+ELAKA+A F
Sbjct 656 NRPIASFIFSGPTGVGKSELAKALAAYYF 684
> At5g50920
Length=929
Score = 94.7 bits (234), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 42/89 (47%), Positives = 63/89 (70%), Gaps = 0/89 (0%)
Query 33 DVAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPK 92
D+ HI+ WTGIP+ K++ DE R+L++ + L R+IGQ +AVKA++ A+ R GL
Sbjct 575 DIQHIVSSWTGIPVEKVSTDESDRLLKMEETLHKRIIGQDEAVKAISRAIRRARVGLKNP 634
Query 93 NKPLGTFMFLGSSGVGKTELAKAVAEEMF 121
N+P+ +F+F G +GVGK+ELAKA+A F
Sbjct 635 NRPIASFIFSGPTGVGKSELAKALAAYYF 663
> SPBC16D10.08c
Length=905
Score = 92.0 bits (227), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 43/89 (48%), Positives = 61/89 (68%), Gaps = 0/89 (0%)
Query 34 VAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPKN 93
+ I+ WTGIP+ ++ E R+L + +LS +VIGQ +AV AVA+A+ + RAGLS N
Sbjct 552 INEIVARWTGIPVTRLKTTEKERLLNMEKVLSKQVIGQNEAVTAVANAIRLSRAGLSDPN 611
Query 94 KPLGTFMFLGSSGVGKTELAKAVAEEMFD 122
+P+ +F+F G SG GKT L KA+A MFD
Sbjct 612 QPIASFLFCGPSGTGKTLLTKALASFMFD 640
> YDR258c
Length=811
Score = 91.7 bits (226), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 46/113 (40%), Positives = 75/113 (66%), Gaps = 7/113 (6%)
Query 16 ALEELTANGRRLALSLHD------VAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVI 69
AL E + +G ++ L LHD ++ ++ TGIP + + + R+L + + L RV+
Sbjct 449 ALSEKSKDGDKVNL-LHDSVTSDDISKVVAKMTGIPTETVMKGDKDRLLYMENSLKERVV 507
Query 70 GQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFD 122
GQ +A+ A++DA+ +QRAGL+ + +P+ +FMFLG +G GKTEL KA+AE +FD
Sbjct 508 GQDEAIAAISDAVRLQRAGLTSEKRPIASFMFLGPTGTGKTELTKALAEFLFD 560
> ECU11g1420
Length=851
Score = 89.7 bits (221), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 47/121 (38%), Positives = 73/121 (60%), Gaps = 6/121 (4%)
Query 2 TNLVPYLVPGRQLAALEELTANGRRLALSLHDVAHILHLWTGIPLGKMTEDEISRVLRLA 61
TN++P + + LE + + L H VA I+ WTGI + ++T E R++ ++
Sbjct 494 TNVIPVIESELKRLELESVV-----VILPSH-VAEIISRWTGIDVKRLTIKENERLMEMS 547
Query 62 DILSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMF 121
+ R+ GQ AV A+ D++ R GL ++P+G+F+ LG +GVGKTELAKAVA E+F
Sbjct 548 SRIKKRIFGQDHAVDAIVDSILQSRVGLDDDDRPVGSFLLLGPTGVGKTELAKAVAMELF 607
Query 122 D 122
D
Sbjct 608 D 608
> SPBC4F6.17c
Length=803
Score = 87.0 bits (214), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 40/95 (42%), Positives = 64/95 (67%), Gaps = 0/95 (0%)
Query 28 ALSLHDVAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRA 87
+++ D+A ++ TGIP + E ++L + + ++IGQ +A+KA+ADA+ + RA
Sbjct 464 SVTSDDIAVVVSRATGIPTTNLMRGERDKLLNMEQTIGKKIIGQDEALKAIADAVRLSRA 523
Query 88 GLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFD 122
GL N+PL +F+FLG +GVGKT L KA+AE +FD
Sbjct 524 GLQNTNRPLASFLFLGPTGVGKTALTKALAEFLFD 558
> At5g51070
Length=945
Score = 80.1 bits (196), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 37/89 (41%), Positives = 57/89 (64%), Gaps = 0/89 (0%)
Query 33 DVAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPK 92
D+A + +W+GIP+ ++T DE ++ L D L RV+GQ +AV A++ A+ R GL
Sbjct 594 DIAAVASVWSGIPVQQITADERMLLMSLEDQLRGRVVGQDEAVAAISRAVKRSRVGLKDP 653
Query 93 NKPLGTFMFLGSSGVGKTELAKAVAEEMF 121
++P+ +F G +GVGKTEL KA+A F
Sbjct 654 DRPIAAMLFCGPTGVGKTELTKALAANYF 682
Score = 27.7 bits (60), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 32/59 (54%), Gaps = 9/59 (15%)
Query 59 RLADILSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVA 117
R ++ L VIG+++ V+ V IQ KN P+ LG +GVGKT +A+ +A
Sbjct 281 RASEGLIDPVIGREKEVQRV-----IQILCRRTKNNPI----LLGEAGVGKTAIAEGLA 330
> YLL026w
Length=908
Score = 74.7 bits (182), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 38/81 (46%), Positives = 58/81 (71%), Gaps = 1/81 (1%)
Query 42 TGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMF 101
TGIP+ K++E E +++ + LSS V+GQ A+KAV++A+ + R+GL+ +P +F+F
Sbjct 554 TGIPVKKLSESENEKLIHMERDLSSEVVGQMDAIKAVSNAVRLSRSGLANPRQP-ASFLF 612
Query 102 LGSSGVGKTELAKAVAEEMFD 122
LG SG GKTELAK VA +F+
Sbjct 613 LGLSGSGKTELAKKVAGFLFN 633
> Hs13540606
Length=707
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 26/57 (45%), Positives = 35/57 (61%), Gaps = 1/57 (1%)
Query 64 LSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEM 120
L +IGQ+ A+ V A+ + G + PL F+FLGSSG+GKTELAK A+ M
Sbjct 343 LKEHIIGQESAIATVGAAIRRKENGWYDEEHPL-VFLFLGSSGIGKTELAKQTAKYM 398
> At2g25030
Length=265
Score = 44.7 bits (104), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 30/48 (62%), Gaps = 0/48 (0%)
Query 60 LADILSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGV 107
L IL R+I Q V++VADA+ +AG+S N+ + +FMF+G V
Sbjct 2 LEQILHERIIAQDLDVESVADAIRCSKAGISDPNRLIASFMFMGQPSV 49
> At5g40090
Length=459
Score = 33.9 bits (76), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 32/57 (56%), Gaps = 6/57 (10%)
Query 65 SSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMF 121
S+ ++ + +K V D LA++ NK + T GS+GVGKT LA+ + E+F
Sbjct 178 SNALVAMDRHMKVVYDLLALE------VNKEVRTIGIWGSAGVGKTTLARYIYAEIF 228
> Hs21396489
Length=959
Score = 33.1 bits (74), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 23/82 (28%), Positives = 40/82 (48%), Gaps = 4/82 (4%)
Query 36 HILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPKNKP 95
+ L T IP GK + + + + R +L G + K + + +A+ + S + K
Sbjct 460 NYLDWLTSIPWGKYSNENLD-LARAQAVLEEDHYGMEDVKKRILEFIAVSQLRGSTQGKI 518
Query 96 LGTFMFLGSSGVGKTELAKAVA 117
L F G GVGKT +A+++A
Sbjct 519 L---CFYGPPGVGKTSIARSIA 537
> HsM4758682
Length=937
Score = 33.1 bits (74), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 23/82 (28%), Positives = 40/82 (48%), Gaps = 4/82 (4%)
Query 36 HILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPKNKP 95
+ L T IP GK + + + + R +L G + K + + +A+ + S + K
Sbjct 438 NYLDWLTSIPWGKYSNENLD-LARAQAVLEEDHYGMEDVKKRILEFIAVSQLRGSTQGKI 496
Query 96 LGTFMFLGSSGVGKTELAKAVA 117
L F G GVGKT +A+++A
Sbjct 497 L---CFYGPPGVGKTSIARSIA 515
> At4g24850
Length=442
Score = 32.7 bits (73), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 35/74 (47%), Gaps = 15/74 (20%)
Query 49 MTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPK---NKPLGTFMFLGSS 105
+T D+I + ++ DIL V+ L +QR L K KP + G
Sbjct 137 VTFDDIGALEKVKDILKELVM------------LPLQRPELFCKGELTKPCKGILLFGPP 184
Query 106 GVGKTELAKAVAEE 119
G GKT LAKAVA+E
Sbjct 185 GTGKTMLAKAVAKE 198
> SPAC4G9.12
Length=193
Score = 32.3 bits (72), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 24/33 (72%), Gaps = 1/33 (3%)
Query 89 LSPKNKPLG-TFMFLGSSGVGKTELAKAVAEEM 120
++P N+P F+ +G +G GKT +AKAV+E++
Sbjct 6 INPTNQPYKYVFVVIGPAGSGKTTMAKAVSEKL 38
> At5g45720
Length=900
Score = 32.3 bits (72), Expect = 0.20, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 30/53 (56%), Gaps = 8/53 (15%)
Query 68 VIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEM 120
++GQ V+A+++A+A +R GL ++F G +G GKT A+ A +
Sbjct 357 LLGQNLVVQALSNAIAKRRVGL--------LYVFHGPNGTGKTSCARVFARAL 401
> At4g02470
Length=371
Score = 32.3 bits (72), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 37/119 (31%), Positives = 49/119 (41%), Gaps = 28/119 (23%)
Query 14 LAALEELTANGRRLALSLHDVAHILHLWTGIPLGKMTEDEISRVLRLADIL-------SS 66
L L ++ + L SL DV +TE+E + L L+D++ S
Sbjct 24 LQTLHDIQNENKSLKKSLKDV--------------VTENEFEKKL-LSDVIPPSDIGVSF 68
Query 67 RVIGQQQAVKAVADALA---IQRAGLSPK---NKPLGTFMFLGSSGVGKTELAKAVAEE 119
IG + VK L +QR L K KP + G G GKT LAKAVA E
Sbjct 69 DDIGALENVKETLKELVMLPLQRPELFDKGQLTKPTKGILLFGPPGTGKTMLAKAVATE 127
> At1g02890_2
Length=563
Score = 31.6 bits (70), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 30/84 (35%), Positives = 40/84 (47%), Gaps = 14/84 (16%)
Query 49 MTEDEISRVLRLADILSSRVIGQQ----QAVKAVADALA------IQRAGLSPK---NKP 95
+TE+E + L L+D++ IG A++ V D L +QR L K KP
Sbjct 241 VTENEFEKKL-LSDVIPPSDIGVSFSDIGALENVKDTLKELVMLPLQRPELFGKGQLTKP 299
Query 96 LGTFMFLGSSGVGKTELAKAVAEE 119
+ G G GKT LAKAVA E
Sbjct 300 TKGILLFGPPGTGKTMLAKAVATE 323
> CE23016
Length=773
Score = 31.2 bits (69), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 39/83 (46%), Gaps = 4/83 (4%)
Query 38 LHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLG 97
L L + +P T D+I + + IL+ + V + LA+ + S K L
Sbjct 275 LELVSSLPWNTSTIDDI-ELHKARTILTESHEAMDDVKERVLEHLAVCKMNNSVKGMIL- 332
Query 98 TFMFLGSSGVGKTELAKAVAEEM 120
F G G+GKT +AKA+AE M
Sbjct 333 --CFTGPPGIGKTSIAKAIAESM 353
> At2g27600
Length=435
Score = 31.2 bits (69), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 21/61 (34%), Positives = 32/61 (52%), Gaps = 3/61 (4%)
Query 62 DILSSRVIGQQQAVKAVADALAIQ---RAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAE 118
+I S V G + A +A+ +A+ + + K +P F+ G G GK+ LAKAVA
Sbjct 128 NIKWSDVAGLESAKQALQEAVILPVKFPQFFTGKRRPWRAFLLYGPPGTGKSYLAKAVAT 187
Query 119 E 119
E
Sbjct 188 E 188
> At1g48900
Length=522
Score = 30.8 bits (68), Expect = 0.59, Method: Composition-based stats.
Identities = 19/69 (27%), Positives = 30/69 (43%), Gaps = 3/69 (4%)
Query 45 PLGKMTEDEISRVLRLADILS---SRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMF 101
PL K + I +++ L D+ + R I +Q + L + +PK MF
Sbjct 47 PLVKEMQSNIKKIVNLEDLAAGHNKRRIIEQAIFSELCKMLDPGKPAFAPKKAKASVVMF 106
Query 102 LGSSGVGKT 110
+G G GKT
Sbjct 107 VGLQGAGKT 115
> At5g46260
Length=1205
Score = 30.4 bits (67), Expect = 0.81, Method: Composition-based stats.
Identities = 22/75 (29%), Positives = 38/75 (50%), Gaps = 3/75 (4%)
Query 47 GKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSG 106
KM E+ + VLR + +S+ + + D +A A L ++K + GSSG
Sbjct 157 AKMIEEIANDVLRKLLLTTSKDF---EDFVGLEDHIANMSALLDLESKEVKMVGIWGSSG 213
Query 107 VGKTELAKAVAEEMF 121
+GKT +A+A+ +F
Sbjct 214 IGKTTIARALFNNLF 228
> At1g62130
Length=372
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 14/26 (53%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 94 KPLGTFMFLGSSGVGKTELAKAVAEE 119
+P + G SG GKT LAKAVA E
Sbjct 135 QPCNGILLFGPSGTGKTMLAKAVATE 160
> 7294649
Length=398
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 16/35 (45%), Positives = 22/35 (62%), Gaps = 4/35 (11%)
Query 86 RAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEM 120
R G+SP P G ++ G G GKT LA+A+A +M
Sbjct 170 RVGISP---PKGCLLY-GPPGTGKTLLARAIASQM 200
> CE29280
Length=406
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 16/36 (44%), Positives = 23/36 (63%), Gaps = 4/36 (11%)
Query 85 QRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEM 120
+R G++P P G +F G G GKT LA+AVA ++
Sbjct 177 KRVGITP---PKGCLLF-GPPGTGKTLLARAVASQL 208
> Hs4505725
Length=1283
Score = 30.0 bits (66), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 97 GTFMFLGSSGVGKTELAKAVAEEMFD 122
G + G G GK+ LAKA+ +E FD
Sbjct 593 GALLLTGGKGSGKSTLAKAICKEAFD 618
> At5g46490
Length=858
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 22/75 (29%), Positives = 37/75 (49%), Gaps = 3/75 (4%)
Query 47 GKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSG 106
KM E+ + VLR + +S+ + D +A A L ++K + GSSG
Sbjct 157 AKMIEEIANDVLRKLLLTTSKDFDD---FVGLEDHIANMSALLDLESKEVKMVGIWGSSG 213
Query 107 VGKTELAKAVAEEMF 121
+GKT +A+A+ +F
Sbjct 214 IGKTTIARALFNNLF 228
> YOR259c
Length=437
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 17/34 (50%), Positives = 20/34 (58%), Gaps = 4/34 (11%)
Query 84 IQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVA 117
QR G+ P P G ++ G G GKT LAKAVA
Sbjct 207 FQRVGIKP---PKGVLLY-GPPGTGKTLLAKAVA 236
> SPAC17A5.01
Length=948
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 14/27 (51%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 94 KPLGTFMFLGSSGVGKTELAKAVAEEM 120
KP + G G GKT LAKAVA E+
Sbjct 686 KPRSGVLLYGPPGTGKTLLAKAVATEL 712
> YPR111w
Length=564
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 25/75 (33%), Positives = 35/75 (46%), Gaps = 16/75 (21%)
Query 54 ISRVLRLADILSSRVIGQQQAVKAVAD-------ALAIQRAG-------LSPKNKPLGTF 99
ISR R +L R + QQ++VK V++ AL +QR L PK+K
Sbjct 115 ISRRQRTKQVL--RYLEQQRSVKNVSNKVLNEEWALYLQREHEVLRKRRLKPKHKDFQIL 172
Query 100 MFLGSSGVGKTELAK 114
+G G G+ LAK
Sbjct 173 TQVGQGGYGQVYLAK 187
> Hs20562018
Length=852
Score = 29.3 bits (64), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 26/87 (29%), Positives = 44/87 (50%), Gaps = 8/87 (9%)
Query 36 HILHLWTGIPLGKMTEDEISRVLRLADIL-SSRVIGQQQAVKAVADALAIQRAGLSPKNK 94
+ L L +P K T D + +R A IL + ++ K V + LA+++ KN
Sbjct 312 NYLELMVELPWNKSTTDRLD--IRAARILLDNDHYAMEKLKKRVLEYLAVRQL----KNN 365
Query 95 PLGTFM-FLGSSGVGKTELAKAVAEEM 120
G + F+G GVGKT + ++VA+ +
Sbjct 366 LKGPILCFVGPPGVGKTSVGRSVAKTL 392
> At3g01610
Length=703
Score = 29.3 bits (64), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 94 KPLGTFMFLGSSGVGKTELAKAVAEE 119
KP +F G G GKT+LA A+A E
Sbjct 148 KPPSGILFHGPPGCGKTKLANAIANE 173
> At3g53230
Length=815
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 16/35 (45%), Positives = 20/35 (57%), Gaps = 4/35 (11%)
Query 85 QRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEE 119
++ G+SP L F G G GKT LAKA+A E
Sbjct 508 EKFGMSPSKGVL----FYGPPGCGKTLLAKAIANE 538
> At3g09840
Length=809
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 16/35 (45%), Positives = 20/35 (57%), Gaps = 4/35 (11%)
Query 85 QRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEE 119
++ G+SP L F G G GKT LAKA+A E
Sbjct 507 EKFGMSPSKGVL----FYGPPGCGKTLLAKAIANE 537
> Hs4506215
Length=389
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 16/36 (44%), Positives = 22/36 (61%), Gaps = 4/36 (11%)
Query 85 QRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEM 120
QR G+ P P G ++ G G GKT LA+AVA ++
Sbjct 160 QRVGIIP---PKGCLLY-GPPGTGKTLLARAVASQL 191
> At5g03340
Length=843
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 16/35 (45%), Positives = 20/35 (57%), Gaps = 4/35 (11%)
Query 85 QRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEE 119
++ G+SP L F G G GKT LAKA+A E
Sbjct 540 EKFGMSPSKGVL----FYGPPGCGKTLLAKAIANE 570
> At4g19500
Length=1239
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 20/73 (27%), Positives = 41/73 (56%), Gaps = 7/73 (9%)
Query 48 KMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGV 107
+M D++S+ L + S ++G + ++A++ L ++ S K + +G G SG+
Sbjct 163 EMVADDVSKKLFKSSNDFSDIVGIEAHLEAMSSILRLK----SEKARMVG---ISGPSGI 215
Query 108 GKTELAKAVAEEM 120
GKT +AKA+ ++
Sbjct 216 GKTTIAKALFSKL 228
> SPAC23D3.02
Length=340
Score = 28.9 bits (63), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 20/77 (25%), Positives = 34/77 (44%), Gaps = 5/77 (6%)
Query 45 PLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGS 104
P K TE E + + ++ + + Q V + + + + L N P +F GS
Sbjct 6 PRNKKTEQEAKKSIPWVELYRPKTLDQ---VSSQESTVQVLKKTLLSNNLP--HMLFYGS 60
Query 105 SGVGKTELAKAVAEEMF 121
G GKT A++ E+F
Sbjct 61 PGTGKTSTILALSRELF 77
> At3g05790
Length=942
Score = 28.9 bits (63), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 31/113 (27%), Positives = 50/113 (44%), Gaps = 9/113 (7%)
Query 9 VPGRQLAALEELTANGRRLALSLHD---VAHILHLWTGIPLGKMTEDEISRVLRLADILS 65
+PG L +EE + L S + + L T +P G + DE VLR IL
Sbjct 363 IPGHVLKVIEEELKKLQLLETSSSEFDVTCNYLDWLTVLPWGNFS-DENFNVLRAEKILD 421
Query 66 SRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFL-GSSGVGKTELAKAVA 117
G + + + +A+ GL + G + L G +GVGKT + +++A
Sbjct 422 EDHYGLSDVKERILEFIAV--GGL--RGTSQGKIICLSGPTGVGKTSIGRSIA 470
> YDL126c
Length=835
Score = 28.9 bits (63), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 17/33 (51%), Positives = 19/33 (57%), Gaps = 4/33 (12%)
Query 88 GLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEM 120
GLSP +F G G GKT LAKAVA E+
Sbjct 517 GLSPSK----GVLFYGPPGTGKTLLAKAVATEV 545
Score = 28.5 bits (62), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 25/88 (28%), Positives = 39/88 (44%), Gaps = 12/88 (13%)
Query 37 ILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVAD-----ALAIQRAGLSP 91
I+H W G P+ + E+ + DI R Q ++ + + + G+ P
Sbjct 191 IIH-WEGEPINREDEENNMNEVGYDDIGGCR--KQMAQIREMVELPLRHPQLFKAIGIKP 247
Query 92 KNKPLGTFMFLGSSGVGKTELAKAVAEE 119
P G M+ G G GKT +A+AVA E
Sbjct 248 ---PRGVLMY-GPPGTGKTLMARAVANE 271
> 7290699
Length=397
Score = 28.5 bits (62), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 22/35 (62%), Gaps = 4/35 (11%)
Query 86 RAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEM 120
R G++P P G ++ G G GKT LA+AVA ++
Sbjct 169 RVGITP---PKGCLLY-GPPGTGKTLLARAVASQL 199
> CE18253
Length=598
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 12/28 (42%), Positives = 18/28 (64%), Gaps = 3/28 (10%)
Query 91 PKNKPLGTFMFLGSSGVGKTELAKAVAE 118
P+NK T + G SG+GKT L + +A+
Sbjct 405 PRNK---TLLITGDSGIGKTSLMRVIAD 429
> At1g64110
Length=821
Score = 28.5 bits (62), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 94 KPLGTFMFLGSSGVGKTELAKAVAEE 119
KP + G G GKT LAKA+A+E
Sbjct 545 KPCRGILLFGPPGTGKTMLAKAIAKE 570
> At1g15310
Length=479
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 17/63 (26%), Positives = 29/63 (46%), Gaps = 3/63 (4%)
Query 51 EDEISRVLRLADILS---SRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGV 107
+ I +++ L D+ + R+I +Q K + L + +PK MF+G G
Sbjct 53 QTNIKKIVNLDDLAAGHNKRLIIEQAIFKELCRMLDPGKPAFAPKKAKPSVVMFVGLQGA 112
Query 108 GKT 110
GKT
Sbjct 113 GKT 115
> SPAC1565.08
Length=418
Score = 28.5 bits (62), Expect = 3.5, Method: Composition-based stats.
Identities = 27/91 (29%), Positives = 45/91 (49%), Gaps = 18/91 (19%)
Query 37 ILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAI--------QRAG 88
I+H W G P+ + EDE S LA++ + G ++ + + + + + + G
Sbjct 201 IIH-WEGEPINR--EDEESS---LAEVGYDDIGGCRRQMAQIRELVELPLRHPQLFKSIG 254
Query 89 LSPKNKPLGTFMFLGSSGVGKTELAKAVAEE 119
+ P P G M+ G G GKT +A+AVA E
Sbjct 255 IKP---PRGILMY-GPPGTGKTLMARAVANE 281
> Hs13027640
Length=926
Score = 28.1 bits (61), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 27/48 (56%), Gaps = 2/48 (4%)
Query 73 QAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEM 120
QAV+AV DA GL PK+ TF+F G+ V K A+A+ E+
Sbjct 193 QAVQAVRDAGYEISLGLMPKSIGPLTFVFTGTGNVSKG--AQAIFNEL 238
Lambda K H
0.319 0.134 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194805952
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40