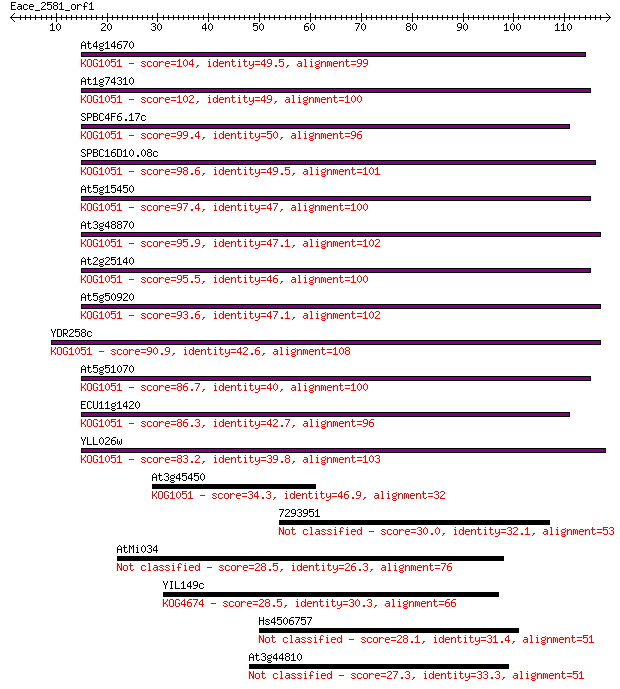
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2581_orf1
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
At4g14670 104 4e-23
At1g74310 102 2e-22
SPBC4F6.17c 99.4 2e-21
SPBC16D10.08c 98.6 2e-21
At5g15450 97.4 6e-21
At3g48870 95.9 2e-20
At2g25140 95.5 2e-20
At5g50920 93.6 8e-20
YDR258c 90.9 5e-19
At5g51070 86.7 1e-17
ECU11g1420 86.3 1e-17
YLL026w 83.2 1e-16
At3g45450 34.3 0.050
7293951 30.0 0.94
AtMi034 28.5 3.1
YIL149c 28.5 3.5
Hs4506757 28.1 4.0
At3g44810 27.3 6.5
> At4g14670
Length=623
Score = 104 bits (259), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 49/99 (49%), Positives = 66/99 (66%), Gaps = 0/99 (0%)
Query 15 ARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKANYEKFHSV 74
ARG++ +GATTLEEY+ H+EKDAAF RRFQ + V PS +SIL+ +K YE H V
Sbjct 270 ARGQLRFIGATTLEEYRTHVEKDAAFERRFQQVFVAEPSVPDTISILRGLKEKYEGHHGV 329
Query 75 DLSDEVLEGIVALSDQYVKQRSFPDKALDLLDEACSWKK 113
+ D L LS++Y+ R PDKA+DL+DE+C+ K
Sbjct 330 RIQDRALVLSAQLSERYITGRRLPDKAIDLVDESCAHVK 368
> At1g74310
Length=911
Score = 102 bits (253), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 49/100 (49%), Positives = 66/100 (66%), Gaps = 0/100 (0%)
Query 15 ARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKANYEKFHSV 74
ARG++ +GATTLEEY+ ++EKDAAF RRFQ + V PS +SIL+ +K YE H V
Sbjct 305 ARGQLRCIGATTLEEYRKYVEKDAAFERRFQQVYVAEPSVPDTISILRGLKEKYEGHHGV 364
Query 75 DLSDEVLEGIVALSDQYVKQRSFPDKALDLLDEACSWKKV 114
+ D L LS +Y+ R PDKA+DL+DEAC+ +V
Sbjct 365 RIQDRALINAAQLSARYITGRHLPDKAIDLVDEACANVRV 404
> SPBC4F6.17c
Length=803
Score = 99.4 bits (246), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 48/96 (50%), Positives = 64/96 (66%), Gaps = 0/96 (0%)
Query 15 ARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKANYEKFHSV 74
ARG++ GATTLEEY+ +IEKDAA RRFQ ++V PS +SIL+ +K YE H V
Sbjct 237 ARGKLHCCGATTLEEYRKYIEKDAALARRFQAVMVNEPSVADTISILRGLKERYEVHHGV 296
Query 75 DLSDEVLEGIVALSDQYVKQRSFPDKALDLLDEACS 110
++D+ L S +Y+ R PDKA+DL+DEACS
Sbjct 297 RITDDALVTAATYSARYITDRFLPDKAIDLVDEACS 332
> SPBC16D10.08c
Length=905
Score = 98.6 bits (244), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 50/101 (49%), Positives = 68/101 (67%), Gaps = 0/101 (0%)
Query 15 ARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKANYEKFHSV 74
ARG++ +GATTL EYK +IEKDAAF RRFQ I+V+ PS E +SIL+ +K YE H V
Sbjct 308 ARGKLHCIGATTLAEYKKYIEKDAAFERRFQIILVKEPSIEDTISILRGLKEKYEVHHGV 367
Query 75 DLSDEVLEGIVALSDQYVKQRSFPDKALDLLDEACSWKKVS 115
+SD L L+ +Y+ R PD A+DL+DEA + +V+
Sbjct 368 TISDRALVTAAHLASRYLTSRRLPDSAIDLVDEAAAAVRVT 408
> At5g15450
Length=968
Score = 97.4 bits (241), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 47/100 (47%), Positives = 66/100 (66%), Gaps = 0/100 (0%)
Query 15 ARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKANYEKFHSV 74
RGE+ +GATTL+EY+ +IEKD A RRFQ + V+ P+ E +SIL+ ++ YE H V
Sbjct 380 GRGELRCIGATTLDEYRKYIEKDPALERRFQQVYVDQPTVEDTISILRGLRERYELHHGV 439
Query 75 DLSDEVLEGIVALSDQYVKQRSFPDKALDLLDEACSWKKV 114
+SD L LSD+Y+ R PDKA+DL+DEA + K+
Sbjct 440 RISDSALVEAAILSDRYISGRFLPDKAIDLVDEAAAKLKM 479
> At3g48870
Length=952
Score = 95.9 bits (237), Expect = 2e-20, Method: Composition-based stats.
Identities = 48/102 (47%), Positives = 66/102 (64%), Gaps = 0/102 (0%)
Query 15 ARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKANYEKFHSV 74
ARGE+ +GATT++EY+ HIEKD A RRFQ + V P+ E A+ IL+ ++ YE H +
Sbjct 420 ARGELQCIGATTIDEYRKHIEKDPALERRFQPVKVPEPTVEEAIQILQGLRERYEIHHKL 479
Query 75 DLSDEVLEGIVALSDQYVKQRSFPDKALDLLDEACSWKKVSH 116
+DE L LS QY+ R PDKA+DL+DEA S ++ H
Sbjct 480 RYTDEALVAAAQLSHQYISDRFLPDKAIDLIDEAGSRVRLRH 521
> At2g25140
Length=874
Score = 95.5 bits (236), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 46/100 (46%), Positives = 65/100 (65%), Gaps = 0/100 (0%)
Query 15 ARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKANYEKFHSV 74
RGE+ +GATTL EY+ +IEKD A RRFQ ++ PS E +SIL+ ++ YE H V
Sbjct 295 GRGELRCIGATTLTEYRKYIEKDPALERRFQQVLCVQPSVEDTISILRGLRERYELHHGV 354
Query 75 DLSDEVLEGIVALSDQYVKQRSFPDKALDLLDEACSWKKV 114
+SD L L+D+Y+ +R PDKA+DL+DEA + K+
Sbjct 355 TISDSALVSAAVLADRYITERFLPDKAIDLVDEAGAKLKM 394
> At5g50920
Length=929
Score = 93.6 bits (231), Expect = 8e-20, Method: Composition-based stats.
Identities = 48/102 (47%), Positives = 65/102 (63%), Gaps = 0/102 (0%)
Query 15 ARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKANYEKFHSV 74
ARGE+ +GATTL+EY+ HIEKD A RRFQ + V P+ + + ILK ++ YE H +
Sbjct 399 ARGELQCIGATTLDEYRKHIEKDPALERRFQPVKVPEPTVDETIQILKGLRERYEIHHKL 458
Query 75 DLSDEVLEGIVALSDQYVKQRSFPDKALDLLDEACSWKKVSH 116
+DE L LS QY+ R PDKA+DL+DEA S ++ H
Sbjct 459 RYTDESLVAAAQLSYQYISDRFLPDKAIDLIDEAGSRVRLRH 500
> YDR258c
Length=811
Score = 90.9 bits (224), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 46/108 (42%), Positives = 69/108 (63%), Gaps = 2/108 (1%)
Query 9 VIGQQQARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKANY 68
++ + ARG + + ATTL+E+K+ IEKD A RRFQ I++ PS +SIL+ +K Y
Sbjct 235 ILKPKLARG-LRCISATTLDEFKI-IEKDPALSRRFQPILLNEPSVSDTISILRGLKERY 292
Query 69 EKFHSVDLSDEVLEGIVALSDQYVKQRSFPDKALDLLDEACSWKKVSH 116
E H V ++D L LS++Y+ R PDKA+DL+DEAC+ ++ H
Sbjct 293 EVHHGVRITDTALVSAAVLSNRYITDRFLPDKAIDLVDEACAVLRLQH 340
> At5g51070
Length=945
Score = 86.7 bits (213), Expect = 1e-17, Method: Composition-based stats.
Identities = 40/100 (40%), Positives = 63/100 (63%), Gaps = 0/100 (0%)
Query 15 ARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKANYEKFHSV 74
RGE+ + +TTL+E++ EKD A RRFQ +++ PS+E A+ IL ++ YE H+
Sbjct 418 GRGELQCIASTTLDEFRSQFEKDKALARRFQPVLINEPSEEDAVKILLGLREKYEAHHNC 477
Query 75 DLSDEVLEGIVALSDQYVKQRSFPDKALDLLDEACSWKKV 114
+ E ++ V LS +Y+ R PDKA+DL+DEA S ++
Sbjct 478 KYTMEAIDAAVYLSSRYIADRFLPDKAIDLIDEAGSRARI 517
> ECU11g1420
Length=851
Score = 86.3 bits (212), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 41/96 (42%), Positives = 60/96 (62%), Gaps = 0/96 (0%)
Query 15 ARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKANYEKFHSV 74
A G I +GATT +EY+ +IE D AF RRF +VV PS E ++++L+ +K E +H V
Sbjct 291 AAGTIKCIGATTHDEYRKYIENDPAFERRFVQVVVNEPSIEDSITMLRGLKGRLEAYHGV 350
Query 75 DLSDEVLEGIVALSDQYVKQRSFPDKALDLLDEACS 110
++D + V S +Y+ R PD A+DLLD AC+
Sbjct 351 KIADSAIVYAVNSSKKYISNRRLPDVAIDLLDTACA 386
> YLL026w
Length=908
Score = 83.2 bits (204), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 41/103 (39%), Positives = 63/103 (61%), Gaps = 0/103 (0%)
Query 15 ARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKANYEKFHSV 74
+RG++ ++GATT EY+ +EKD AF RRFQ I V PS + ++IL+ ++ YE H V
Sbjct 306 SRGQLKVIGATTNNEYRSIVEKDGAFERRFQKIEVAEPSVRQTVAILRGLQPKYEIHHGV 365
Query 75 DLSDEVLEGIVALSDQYVKQRSFPDKALDLLDEACSWKKVSHN 117
+ D L L+ +Y+ R PD ALDL+D +C+ V+ +
Sbjct 366 RILDSALVTAAQLAKRYLPYRRLPDSALDLVDISCAGVAVARD 408
> At3g45450
Length=341
Score = 34.3 bits (77), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 29 EYKLHIEKDAAFCRRFQNIVVEAPSKERALSI 60
+Y+ HIE D A RRFQ + V P+ E A+ I
Sbjct 278 QYRKHIENDPALERRFQPVKVPEPTVEEAIQI 309
> 7293951
Length=468
Score = 30.0 bits (66), Expect = 0.94, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 29/53 (54%), Gaps = 0/53 (0%)
Query 54 KERALSILKKVKANYEKFHSVDLSDEVLEGIVALSDQYVKQRSFPDKALDLLD 106
K + L++L+ + +YE F V + EVL +A + + S PD+ +LLD
Sbjct 83 KVKDLNLLEVTEPHYETFRCVRSNKEVLPDDLADQLKTIADMSIPDRQAELLD 135
> AtMi034
Length=106
Score = 28.5 bits (62), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 20/77 (25%), Positives = 37/77 (48%), Gaps = 2/77 (2%)
Query 22 VGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSK-ERALSILKKVKANYEKFHSVDLSDEV 80
+G T L EY L A+ R + + +K ER+ + +++ + FHS+ + +E
Sbjct 13 MGVTVLPEY-LKQSSYEAYSRPYSAFFLSGCTKQERSPLLARRLVDAWLSFHSILMINEE 71
Query 81 LEGIVALSDQYVKQRSF 97
+ LSD Y ++ F
Sbjct 72 VSDWEQLSDHYTRRSLF 88
> YIL149c
Length=1679
Score = 28.5 bits (62), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 28/66 (42%), Gaps = 9/66 (13%)
Query 31 KLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKANYEKFHSVDLSDEVLEGIVALSDQ 90
KL +EK C+R QNIV++ +E A V K S I L Q
Sbjct 323 KLRLEKSKNECQRLQNIVMDCTKEEEATMTTSAVSPTVGKLFS---------DIKVLKRQ 373
Query 91 YVKQRS 96
+K+R+
Sbjct 374 LIKERN 379
> Hs4506757
Length=4967
Score = 28.1 bits (61), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 16/58 (27%), Positives = 29/58 (50%), Gaps = 7/58 (12%)
Query 50 EAPSKERALSILKKVKANYEKFHSVDLSDEVL-------EGIVALSDQYVKQRSFPDK 100
E PS+ + +L++ K +Y HS L+++VL + ++ S Y R FP +
Sbjct 1379 EKPSRLKQRFLLRRTKPDYSTSHSARLTEDVLADDRDDYDFLMQTSTYYYSVRIFPGQ 1436
> At3g44810
Length=448
Score = 27.3 bits (59), Expect = 6.5, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 27/51 (52%), Gaps = 1/51 (1%)
Query 48 VVEAPSKERALSILKKVKANYEKFHSVDLSDEVLEGIVALSDQYVKQRSFP 98
+V PS + L +LK + + F + S+ +L G AL D + Q+SFP
Sbjct 155 IVYFPS-DTCLPVLKILVLDSIWFDRIKFSNVLLAGCPALEDLTIDQKSFP 204
Lambda K H
0.317 0.132 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167969826
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40