bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
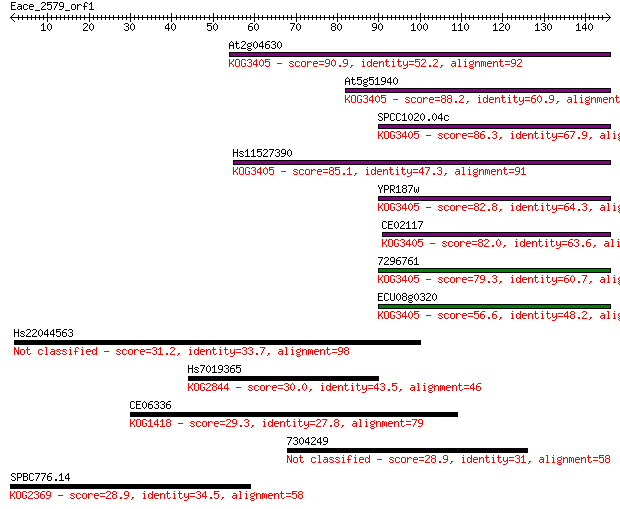
Query= Eace_2579_orf1
Length=145
Score E
Sequences producing significant alignments: (Bits) Value
At2g04630 90.9 8e-19
At5g51940 88.2 4e-18
SPCC1020.04c 86.3 2e-17
Hs11527390 85.1 4e-17
YPR187w 82.8 2e-16
CE02117 82.0 4e-16
7296761 79.3 2e-15
ECU08g0320 56.6 1e-08
Hs22044563 31.2 0.77
Hs7019365 30.0 1.6
CE06336 29.3 2.9
7304249 28.9 3.5
SPBC776.14 28.9 3.6
> At2g04630
Length=144
Score = 90.9 bits (224), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 48/92 (52%), Positives = 65/92 (70%), Gaps = 7/92 (7%)
Query 54 DEDIEEADAAAEDVDVVIDPNDPRLQRPEAHPSEGPRITTPFLTKFEKARIIGTRALQIS 113
D DI+E D DV +DP + + E P + PR T+ F+TK+E+ARI+GTRALQIS
Sbjct 30 DADIKEND------DVNVDPLETE-DKVETEPVQRPRKTSKFMTKYERARILGTRALQIS 82
Query 114 MNAPLTIQLEGESDPLIIAEKELYQKTLPFIV 145
MNAP+ ++LEGE+DPL IA KEL Q+ +PF +
Sbjct 83 MNAPVMVELEGETDPLEIAMKELRQRKIPFTI 114
> At5g51940
Length=144
Score = 88.2 bits (217), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 39/64 (60%), Positives = 52/64 (81%), Gaps = 0/64 (0%)
Query 82 EAHPSEGPRITTPFLTKFEKARIIGTRALQISMNAPLTIQLEGESDPLIIAEKELYQKTL 141
E P + PR T+ F+TK+E+ARI+GTRALQISMNAP+ ++LEGE+DPL IA KEL Q+ +
Sbjct 51 ETEPVQRPRKTSKFMTKYERARILGTRALQISMNAPVMVELEGETDPLEIAMKELRQRKI 110
Query 142 PFIV 145
PF +
Sbjct 111 PFTI 114
> SPCC1020.04c
Length=142
Score = 86.3 bits (212), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 38/56 (67%), Positives = 48/56 (85%), Gaps = 0/56 (0%)
Query 90 RITTPFLTKFEKARIIGTRALQISMNAPLTIQLEGESDPLIIAEKELYQKTLPFIV 145
R TTP++TK+E+ARI+GTRALQISMNAP+ + LEGE+DPL IA KEL QK +P +V
Sbjct 69 RTTTPYMTKYERARILGTRALQISMNAPVLVDLEGETDPLQIAMKELAQKKIPLLV 124
> Hs11527390
Length=127
Score = 85.1 bits (209), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 43/92 (46%), Positives = 68/92 (73%), Gaps = 9/92 (9%)
Query 55 EDIEEADA-AAEDVDVVIDPNDPRLQRPEAHPSEGPRITTPFLTKFEKARIIGTRALQIS 113
+D+E A+ E+V+++ P +RP+A+ RITTP++TK+E+AR++GTRALQI+
Sbjct 23 DDLENAEEEGQENVEIL-----PSGERPQANQK---RITTPYMTKYERARVLGTRALQIA 74
Query 114 MNAPLTIQLEGESDPLIIAEKELYQKTLPFIV 145
M AP+ ++LEGE+DPL+IA KEL + +P I+
Sbjct 75 MCAPVMVELEGETDPLLIAMKELKARKIPIII 106
> YPR187w
Length=155
Score = 82.8 bits (203), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 36/56 (64%), Positives = 48/56 (85%), Gaps = 0/56 (0%)
Query 90 RITTPFLTKFEKARIIGTRALQISMNAPLTIQLEGESDPLIIAEKELYQKTLPFIV 145
R TTP++TK+E+ARI+GTRALQISMNAP+ + LEGE+DPL IA KEL +K +P ++
Sbjct 79 RATTPYMTKYERARILGTRALQISMNAPVFVDLEGETDPLRIAMKELAEKKIPLVI 134
> CE02117
Length=137
Score = 82.0 bits (201), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 35/55 (63%), Positives = 48/55 (87%), Gaps = 0/55 (0%)
Query 91 ITTPFLTKFEKARIIGTRALQISMNAPLTIQLEGESDPLIIAEKELYQKTLPFIV 145
+TTPF+TK+E+AR++GTRALQI+M AP+ ++LEGE+DPL IA KEL Q+ +P IV
Sbjct 61 VTTPFMTKYERARVLGTRALQIAMGAPVMVELEGETDPLEIARKELKQRRIPIIV 115
> 7296761
Length=131
Score = 79.3 bits (194), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 34/56 (60%), Positives = 48/56 (85%), Gaps = 0/56 (0%)
Query 90 RITTPFLTKFEKARIIGTRALQISMNAPLTIQLEGESDPLIIAEKELYQKTLPFIV 145
RITT ++TK+E+AR++GTRALQI+M AP+ ++L+GE+DPL IA KEL QK +P I+
Sbjct 54 RITTKYMTKYERARVLGTRALQIAMCAPIMVELDGETDPLQIAMKELKQKKIPIII 109
> ECU08g0320
Length=144
Score = 56.6 bits (135), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 27/56 (48%), Positives = 38/56 (67%), Gaps = 0/56 (0%)
Query 90 RITTPFLTKFEKARIIGTRALQISMNAPLTIQLEGESDPLIIAEKELYQKTLPFIV 145
R T P +TK+EKA +IG RA Q+S AP + + E++PL IA EL +K +PFI+
Sbjct 64 RKTLPRMTKYEKAHVIGVRATQLSAGAPPFVDIGNETNPLNIANMELSEKKIPFII 119
> Hs22044563
Length=194
Score = 31.2 bits (69), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 33/106 (31%), Positives = 45/106 (42%), Gaps = 16/106 (15%)
Query 2 STRNVQNKTQKN-----RKNTGLG---FRVLKMADDENDHMFAEPAFGDEFGDELGEEDL 53
STRN+Q Q + RK L F +E DHM +EP G EFG LG +
Sbjct 44 STRNIQFHEQASEMGNFRKGLPLADQFFDYHNQEMEEGDHM-SEPRLGKEFGVNLGGQIG 102
Query 54 DEDIEEADAAAEDVDVVIDPNDPRLQRPEAHPSEGPRITTPFLTKF 99
D + + + D +I D E H E ++T P L+ F
Sbjct 103 DAIVYQKNKQERKADHLISQAD-----QEGHILE--QVTQPGLSDF 141
> Hs7019365
Length=866
Score = 30.0 bits (66), Expect = 1.6, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 24/52 (46%), Gaps = 6/52 (11%)
Query 44 FGDELGEEDLDEDIEEADAAAEDV------DVVIDPNDPRLQRPEAHPSEGP 89
FG EL E DLD +E AA E V D++ N P P+ P GP
Sbjct 333 FGKELFESDLDRIMEHIKAAMEMVPVLKKADIINVVNGPITYSPDILPMVGP 384
> CE06336
Length=522
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 22/79 (27%), Positives = 35/79 (44%), Gaps = 13/79 (16%)
Query 30 ENDHMFAEPAFGDEFGDELGEEDLDEDIEEADAAAEDVDVVIDPNDPRLQRPEAHPSEGP 89
EN +M+A ++ ++L ++ DED ++AD ND + E P GP
Sbjct 314 ENQYMWALELIDQKYQEKLKQDMYDEDEKKADK-----------NDMHFSKKE--PVRGP 360
Query 90 RITTPFLTKFEKARIIGTR 108
RI L + +I G R
Sbjct 361 RILLQDLLRGPDLKISGGR 379
> 7304249
Length=706
Score = 28.9 bits (63), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 18/64 (28%), Positives = 28/64 (43%), Gaps = 6/64 (9%)
Query 68 DVVIDPNDPRLQRPEAHPSEGPRITTPFLTKFEKARIIGTRAL------QISMNAPLTIQ 121
+ +++P RL R P + T + F+K+ GTR L Q +M P +
Sbjct 403 EFILEPQGRRLIRVMFQPKKVGLFTQRWHVNFQKSPFCGTRRLDVVLQGQCTMPVPFQRR 462
Query 122 LEGE 125
LEG
Sbjct 463 LEGH 466
> SPBC776.14
Length=623
Score = 28.9 bits (63), Expect = 3.6, Method: Composition-based stats.
Identities = 20/66 (30%), Positives = 36/66 (54%), Gaps = 8/66 (12%)
Query 1 TSTRNVQNKTQKNRKNTGLGF---RVLKMADD-----ENDHMFAEPAFGDEFGDELGEED 52
+ T++V NK++K++ L F +L + +D+ +PA D+FG+ LG D
Sbjct 39 SETQSVSNKSRKSKFGKRLNFILGAILGICGAFFFAVGDDNAVFDPATLDKFGNMLGSSD 98
Query 53 LDEDIE 58
L +DI+
Sbjct 99 LFDDIK 104
Lambda K H
0.313 0.134 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1712413322
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40