bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2571_orf1
Length=133
Score E
Sequences producing significant alignments: (Bits) Value
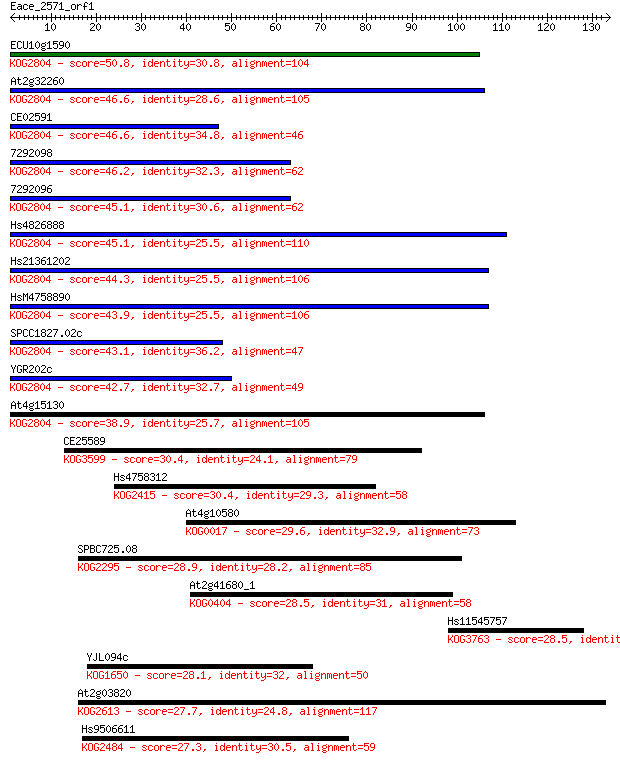
ECU10g1590 50.8 7e-07
At2g32260 46.6 1e-05
CE02591 46.6 1e-05
7292098 46.2 2e-05
7292096 45.1 4e-05
Hs4826888 45.1 4e-05
Hs21361202 44.3 7e-05
HsM4758890 43.9 7e-05
SPCC1827.02c 43.1 1e-04
YGR202c 42.7 2e-04
At4g15130 38.9 0.003
CE25589 30.4 0.92
Hs4758312 30.4 1.0
At4g10580 29.6 1.5
SPBC725.08 28.9 2.9
At2g41680_1 28.5 3.9
Hs11545757 28.5 4.1
YJL094c 28.1 4.6
At2g03820 27.7 6.7
Hs9506611 27.3 7.3
> ECU10g1590
Length=278
Score = 50.8 bits (120), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 32/104 (30%), Positives = 49/104 (47%), Gaps = 16/104 (15%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKV 60
ST+ II I++NY+ YV R+L RGV +D+NI + + IKM K M +E+ K
Sbjct 167 STSGIITSIVKNYDMYVRRNLERGVSAKDLNISFLQMKRIKMSKKM--------NEMIK- 217
Query 61 TLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGFASKF 104
V ++ +R I W S+ ++ GF F
Sbjct 218 -------NVDIQREIDDIRREIRIAMRYWERVSNEIISGFIENF 254
> At2g32260
Length=332
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 30/105 (28%), Positives = 54/105 (51%), Gaps = 15/105 (14%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKV 60
ST+DII+RI+++Y YV R+L RG D+ + + K +++ +++ E+V ++ +V
Sbjct 162 STSDIIMRIVKDYNQYVMRNLDRGYSREDLGVSFVKEKRLRVNMRLKKLQERVKEQQERV 221
Query 61 TLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGFASKFE 105
G ++V+ LRN W + R + GF FE
Sbjct 222 -------GEKI-QTVKMLRNE-------WVENADRWVAGFLEIFE 251
> CE02591
Length=347
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 34/46 (73%), Gaps = 0/46 (0%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNM 46
ST+D++ RI+++Y+ YV R+L RG P+++N+G+ A+ +++ +
Sbjct 190 STSDVVCRIIRDYDKYVRRNLQRGYSPKELNVGFLAASKYQIQNKV 235
> 7292098
Length=381
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 38/62 (61%), Gaps = 0/62 (0%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKV 60
ST+DI+ RI+++Y+ YV R+L+RG +++N+ + +++ M + ELTKV
Sbjct 203 STSDIVARIVKDYDVYVRRNLARGYSAKELNVSFLSEKKFRLQNKMDELKTRGKRELTKV 262
Query 61 TL 62
+
Sbjct 263 KV 264
> 7292096
Length=347
Score = 45.1 bits (105), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 38/62 (61%), Gaps = 0/62 (0%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKV 60
ST+DI+ RI+++Y+ YV R+L+RG +++N+ + +++ M + EL+KV
Sbjct 151 STSDIVARIVKDYDLYVRRNLARGYSAKELNVSFLSEKKFRLQNKMDELKSRGKRELSKV 210
Query 61 TL 62
+
Sbjct 211 KV 212
> Hs4826888
Length=367
Score = 45.1 bits (105), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 28/110 (25%), Positives = 49/110 (44%), Gaps = 11/110 (10%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKV 60
ST+DII RI+++Y+ Y R+L RG +++N+ + +++ + + EKV D K
Sbjct 201 STSDIITRIVRDYDVYARRNLQRGYTAKELNVSFINEKKYHLQERVDKVKEKVKDVEEKS 260
Query 61 TLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGFASKFEPMKSL 110
E V+K+ W +S + F F P +L
Sbjct 261 K-----------EFVQKVEEKSIDLIQKWEEKSREFIGSFLEMFGPEGAL 299
> Hs21361202
Length=369
Score = 44.3 bits (103), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 27/108 (25%), Positives = 48/108 (44%), Gaps = 15/108 (13%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVS--DELT 58
ST+DII RI+++Y+ Y R+L RG +++N+ + + + + + EKV +E +
Sbjct 201 STSDIITRIVRDYDVYARRNLQRGYTAKELNVSFINEKRYRFQNQVDKMKEKVKNVEERS 260
Query 59 KVTLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGFASKFEP 106
K E V ++ H W +S + F F P
Sbjct 261 K-------------EFVNRVEEKSHDLIQKWEEKSREFIGNFLELFGP 295
> HsM4758890
Length=330
Score = 43.9 bits (102), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 27/108 (25%), Positives = 48/108 (44%), Gaps = 15/108 (13%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVS--DELT 58
ST+DII RI+++Y+ Y R+L RG +++N+ + + + + + EKV +E +
Sbjct 201 STSDIITRIVRDYDVYARRNLQRGYTAKELNVSFINEKRYRFQNQVDKMKEKVKNVEERS 260
Query 59 KVTLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGFASKFEP 106
K E V ++ H W +S + F F P
Sbjct 261 K-------------EFVNRVEEKSHDLIQKWEEKSREFIGNFLELFGP 295
> SPCC1827.02c
Length=354
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 35/47 (74%), Gaps = 0/47 (0%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQ 47
ST+D+I RI+++Y+ YV R+L+RGV +++N+ K N + ++ +++
Sbjct 218 STSDLITRIIRDYDQYVMRNLARGVNRKELNVSLFKKNELDLRHHIK 264
> YGR202c
Length=424
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 35/49 (71%), Gaps = 0/49 (0%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRW 49
ST+DII +I+++Y+ Y+ R+ +RG +++N+ + K N ++ KK++ +
Sbjct 228 STSDIITKIIRDYDKYLMRNFARGATRQELNVSWLKKNELEFKKHINEF 276
> At4g15130
Length=298
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 27/105 (25%), Positives = 48/105 (45%), Gaps = 17/105 (16%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKV 60
ST+DII+RI+++Y YV R+ +++ K + M+ +++ EKV ++ K+
Sbjct 147 STSDIIMRIVKDYNQYVLRNPGPRYSREELSCQLEKRLRVNMR--LKKLQEKVKEQQEKI 204
Query 61 TLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGFASKFE 105
+ G+H DE W + R + GF FE
Sbjct 205 QTVAKTAGMHHDE---------------WLENADRWVAGFLEMFE 234
> CE25589
Length=815
Score = 30.4 bits (67), Expect = 0.92, Method: Composition-based stats.
Identities = 19/86 (22%), Positives = 40/86 (46%), Gaps = 7/86 (8%)
Query 13 YEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNM-QRWGEKVSDELTKVTLTDRPLG--- 68
YEDY G +D+N +T+ N M +++ ++ E ++DE+ ++T R
Sbjct 677 YEDYKLMLYRAGYAEKDINEAFTRFNVTSMTEHVPEKVAEDIADEVARMTEQKRNYMENH 736
Query 69 ---VHFDESVEKLRNSIHQTYDNWRG 91
+ + V++++ S+ D G
Sbjct 737 RDYANLNRRVDQMQESVFSIVDRIEG 762
> Hs4758312
Length=617
Score = 30.4 bits (67), Expect = 1.0, Method: Composition-based stats.
Identities = 17/58 (29%), Positives = 27/58 (46%), Gaps = 1/58 (1%)
Query 24 GVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKVTLTDRPLGVHFDESVEKLRNS 81
G P MN+ K MK + E + ++LT L + +G+H E + L+NS
Sbjct 386 GCSPGFMNVPKIKGTHTAMKSGILA-AESIFNQLTSENLQSKTIGLHVTEYEDNLKNS 442
> At4g10580
Length=1240
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 24/92 (26%), Positives = 42/92 (45%), Gaps = 19/92 (20%)
Query 40 IKMKKNMQRWGE-KVSDELTKVTLTDRPLG----------VHFD-ESVEKLRNSIHQT-- 85
+ +++ M++ E K+ +L+K + R +G V D E +E +R+ T
Sbjct 659 VHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNA 718
Query 86 -----YDNWRGRSHRLLRGFASKFEPMKSLIG 112
+ W G R ++GFAS +PM L G
Sbjct 719 TEIRSFLGWAGYYRRFVKGFASMAQPMTKLTG 750
> SPBC725.08
Length=609
Score = 28.9 bits (63), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 24/89 (26%), Positives = 40/89 (44%), Gaps = 20/89 (22%)
Query 16 YVDRSLSRGVKP---RDMNIGYTKANAIKMKKNMQRWGEKVSDELTKVTLTDRPLGVHFD 72
Y DR+ + G P + + YT+ NA+ ++ + RW K + H
Sbjct 52 YDDRTRASGPYPTFTKPLIDPYTQTNAVSYERFI-RWYSKEN---------------HIS 95
Query 73 ESVEKLRNSIHQTYDNWRGRSH-RLLRGF 100
+ E L NS+H TY+N++ + R R F
Sbjct 96 ATTEDLYNSLHGTYNNYKQDLYARTARSF 124
> At2g41680_1
Length=396
Score = 28.5 bits (62), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 29/59 (49%), Gaps = 6/59 (10%)
Query 41 KMKKNMQRWGEKVSDE-LTKVTLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLR 98
KM+K +RWG ++ E + +++T P V E K + I+ T G + R LR
Sbjct 149 KMRKQAERWGAELYPEDVESLSVTTAPFTVQTSERKVKCHSIIYAT-----GATARRLR 202
> Hs11545757
Length=531
Score = 28.5 bits (62), Expect = 4.1, Method: Composition-based stats.
Identities = 13/30 (43%), Positives = 19/30 (63%), Gaps = 2/30 (6%)
Query 98 RGFASKFEPMKSLIGIHVNHHQSSDDDAAI 127
R F+S+ EP+ G+H + HQ D DAA+
Sbjct 28 RRFSSRSEPVNP--GMHSSSHQQQDGDAAM 55
> YJL094c
Length=873
Score = 28.1 bits (61), Expect = 4.6, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 25/50 (50%), Gaps = 3/50 (6%)
Query 18 DRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKVTLTDRPL 67
+ SLSR D + K+N K+KK + W + V D T +++ D L
Sbjct 517 ETSLSRMTTATDSTL---KSNTFKIKKMVHIWSKSVDDVDTNLSVIDEKL 563
> At2g03820
Length=516
Score = 27.7 bits (60), Expect = 6.7, Method: Composition-based stats.
Identities = 29/129 (22%), Positives = 51/129 (39%), Gaps = 12/129 (9%)
Query 16 YVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKVTLTDRPLGVHFDESV 75
YV L +KP D +GY A M+++ V + L + L + +
Sbjct 357 YVQTHLGHILKPGDQALGYDIYGANVNDNEMEKYRLSVKNGLPEAILIKKCYEEQRERKQ 416
Query 76 EKLRN----SIHQTYDNWRGR-----SHRLLRGFASKFEPMKSL---IGIHVNHHQSSDD 123
+K RN S+ D+ RGR + + F E L I ++ + + +
Sbjct 417 KKSRNWKLKSLPMEMDDSRGRVDPEKTDKEYEEFLRDLEENPELRFNISLYRDKDYQASE 476
Query 124 DAAISDGSG 132
A+++DG G
Sbjct 477 TASMTDGEG 485
> Hs9506611
Length=582
Score = 27.3 bits (59), Expect = 7.3, Method: Composition-based stats.
Identities = 18/61 (29%), Positives = 33/61 (54%), Gaps = 2/61 (3%)
Query 17 VDRS--LSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKVTLTDRPLGVHFDES 74
VDR L R ++ + G A+A+K KK MQ+ +K++ +L+ ++ L + D+
Sbjct 520 VDRRSVLQRIMETDPLQQGQALASALKNKKKMQKRADKIASKLSDSMMSALDLSGNADDG 579
Query 75 V 75
V
Sbjct 580 V 580
Lambda K H
0.316 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1356426142
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40