bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2568_orf1
Length=180
Score E
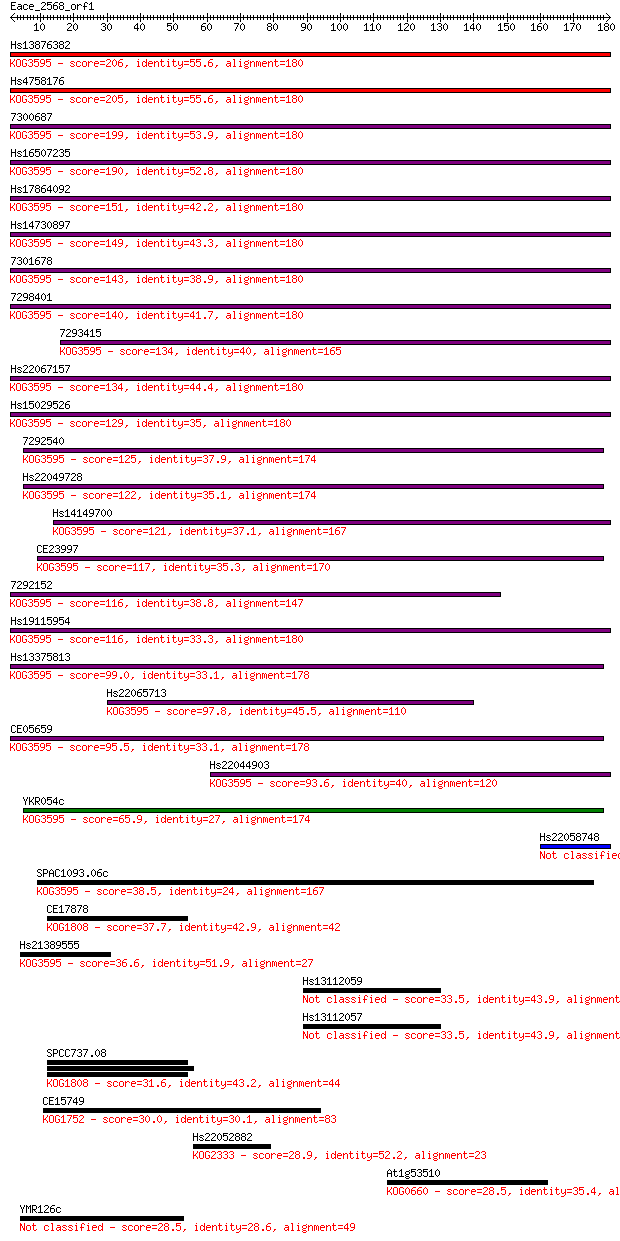
Sequences producing significant alignments: (Bits) Value
Hs13876382 206 2e-53
Hs4758176 205 3e-53
7300687 199 2e-51
Hs16507235 190 1e-48
Hs17864092 151 6e-37
Hs14730897 149 4e-36
7301678 143 1e-34
7298401 140 2e-33
7293415 134 1e-31
Hs22067157 134 1e-31
Hs15029526 129 4e-30
7292540 125 5e-29
Hs22049728 122 3e-28
Hs14149700 121 9e-28
CE23997 117 1e-26
7292152 116 3e-26
Hs19115954 116 3e-26
Hs13375813 99.0 4e-21
Hs22065713 97.8 1e-20
CE05659 95.5 5e-20
Hs22044903 93.6 2e-19
YKR054c 65.9 4e-11
Hs22058748 43.9 2e-04
SPAC1093.06c 38.5 0.007
CE17878 37.7 0.014
Hs21389555 36.6 0.025
Hs13112059 33.5 0.22
Hs13112057 33.5 0.22
SPCC737.08 31.6 0.96
CE15749 30.0 2.9
Hs22052882 28.9 5.2
At1g53510 28.5 8.0
YMR126c 28.5 8.2
> Hs13876382
Length=4486
Score = 206 bits (524), Expect = 2e-53, Method: Compositional matrix adjust.
Identities = 100/187 (53%), Positives = 132/187 (70%), Gaps = 11/187 (5%)
Query 1 QDVVAMQKLDLAHKEGHWVLLQNIHLMPRWTVELEKKLDAFSSEGPHPNFRCFLSSELC- 59
Q+VVA LDLA K+GHWV+LQNIHL+ +W LEKKL+ S E HP FR F+S+E
Sbjct 3938 QEVVAEAALDLAAKKGHWVILQNIHLVAKWLSTLEKKLEEHS-ENSHPEFRVFMSAEPAP 3996
Query 60 ----DYIPVGILDRSIKLTNEPPQGLQANLKRAFACFPKDDFD--EKDQKVKAIIFGLCF 113
IP GIL+ SIK+TNEPP G+ ANL +A F +D + ++ + K+I+F LC+
Sbjct 3997 SPEGHIIPQGILENSIKITNEPPTGMHANLHKALDNFTQDTLEMCSRETEFKSILFALCY 4056
Query 114 FHAVLLERKRFGPRGWNMNYPFAMGDLRDSAMVLFNYMEQQQGGSKVPWDDLKYIFGEIM 173
FHAV+ ER++FGP+GWN +YPF GDL S VL+N++E +KVP+DDL+Y+FGEIM
Sbjct 4057 FHAVVAERRKFGPQGWNRSYPFNTGDLTISVNVLYNFLE---ANAKVPYDDLRYLFGEIM 4113
Query 174 YGGHIID 180
YGGHI D
Sbjct 4114 YGGHITD 4120
> Hs4758176
Length=798
Score = 205 bits (522), Expect = 3e-53, Method: Compositional matrix adjust.
Identities = 100/187 (53%), Positives = 132/187 (70%), Gaps = 11/187 (5%)
Query 1 QDVVAMQKLDLAHKEGHWVLLQNIHLMPRWTVELEKKLDAFSSEGPHPNFRCFLSSELC- 59
Q+VVA LDLA K+GHWV+LQNIHL+ +W LEKKL+ SE HP FR F+S+E
Sbjct 250 QEVVAEAALDLAAKKGHWVILQNIHLVAKWLSTLEKKLEE-HSENSHPEFRVFMSAEPAP 308
Query 60 ----DYIPVGILDRSIKLTNEPPQGLQANLKRAFACFPKDDFD--EKDQKVKAIIFGLCF 113
IP GIL+ SIK+TNEPP G+ ANL +A F +D + ++ + K+I+F LC+
Sbjct 309 SPEGHIIPQGILENSIKITNEPPTGMHANLHKALDNFTQDTLEMCSRETEFKSILFALCY 368
Query 114 FHAVLLERKRFGPRGWNMNYPFAMGDLRDSAMVLFNYMEQQQGGSKVPWDDLKYIFGEIM 173
FHAV+ ER++FGP+GWN +YPF GDL S VL+N++E +KVP+DDL+Y+FGEIM
Sbjct 369 FHAVVAERRKFGPQGWNRSYPFNTGDLTISVNVLYNFLE---ANAKVPYDDLRYLFGEIM 425
Query 174 YGGHIID 180
YGGHI D
Sbjct 426 YGGHITD 432
> 7300687
Length=4472
Score = 199 bits (507), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 97/187 (51%), Positives = 131/187 (70%), Gaps = 11/187 (5%)
Query 1 QDVVAMQKLDLAHKEGHWVLLQNIHLMPRWTVELEKKLDAFSSEGPHPNFRCFLSSELCD 60
Q+ +A +D A K GHWV+LQNIHL+ +W LEKKL+ ++ E HP++R FLS+E
Sbjct 3923 QEAIAEAAMDTAAKHGHWVVLQNIHLVRKWLPVLEKKLEYYA-EDSHPDYRMFLSAEPAS 3981
Query 61 Y-----IPVGILDRSIKLTNEPPQGLQANLKRAFACFPKDDFDE--KDQKVKAIIFGLCF 113
IP GIL+ SIK+TNEPP G+ ANL +A F ++ + K+ + KAI+F LC+
Sbjct 3982 TPSAHIIPQGILESSIKITNEPPTGMLANLHKALDNFTQETLEMSGKEAEFKAILFSLCY 4041
Query 114 FHAVLLERKRFGPRGWNMNYPFAMGDLRDSAMVLFNYMEQQQGGSKVPWDDLKYIFGEIM 173
FHAV+ ER++FGP+GWN YPF +GDL S VL+NY+E +KVPW+DL+Y+FGEIM
Sbjct 4042 FHAVVAERRKFGPQGWNKIYPFNVGDLNISVSVLYNYLE---ANAKVPWEDLRYLFGEIM 4098
Query 174 YGGHIID 180
YGGHI D
Sbjct 4099 YGGHITD 4105
> Hs16507235
Length=4523
Score = 190 bits (482), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 95/187 (50%), Positives = 127/187 (67%), Gaps = 11/187 (5%)
Query 1 QDVVAMQKLDLAHKEGHWVLLQNIHLMPRWTVELEKKLDAFSSEGPHPNFRCFLSSELC- 59
Q+ VA L+ A K GHWV+LQN+HL+ +W LEK L+ FS +G H ++R F+S+E
Sbjct 3975 QETVAEVALEKASKGGHWVILQNVHLVAKWLGTLEKLLERFS-QGSHRDYRVFMSAESAP 4033
Query 60 ----DYIPVGILDRSIKLTNEPPQGLQANLKRAFACFPKDDFD--EKDQKVKAIIFGLCF 113
IP G+L+ SIK+TNEPP G+ ANL A F +D + K+Q+ K+I+F LC+
Sbjct 4034 TPDEHIIPQGLLENSIKITNEPPTGMLANLHAALYNFDQDTLEICSKEQEFKSILFSLCY 4093
Query 114 FHAVLLERKRFGPRGWNMNYPFAMGDLRDSAMVLFNYMEQQQGGSKVPWDDLKYIFGEIM 173
FHA + R RFGP+GW+ +YPF GDL A VL+NY+E SKVPW+DL+Y+FGEIM
Sbjct 4094 FHACVAGRLRFGPQGWSRSYPFNPGDLTICASVLYNYLE---ANSKVPWEDLRYLFGEIM 4150
Query 174 YGGHIID 180
YGGHI D
Sbjct 4151 YGGHITD 4157
> Hs17864092
Length=4024
Score = 151 bits (382), Expect = 6e-37, Method: Compositional matrix adjust.
Identities = 76/185 (41%), Positives = 114/185 (61%), Gaps = 8/185 (4%)
Query 1 QDVVAMQKLDLAHKEGHWVLLQNIHLMPRWTVELEKKLDAFSSEGPHPNFRCFLSSELCD 60
Q +AM+ L+ A KEG WV+LQN HL W LEK + S E HP+FR +L+S
Sbjct 3467 QGPIAMKMLEKAVKEGTWVVLQNCHLATSWMPTLEKVCEELSPESTHPDFRMWLTSYPSP 3526
Query 61 YIPVGILDRSIKLTNEPPQGLQANLKRAFACFPKDDFD-----EKDQKVKAIIFGLCFFH 115
PV +L +K+TNE P+GL+AN+ R++ P D + +K ++ K +++GLCFFH
Sbjct 3527 NFPVSVLQNGVKMTNEAPKGLRANIIRSYLMDPISDPEFFGSCKKPEEFKKLLYGLCFFH 3586
Query 116 AVLLERKRFGPRGWNMNYPFAMGDLRDSAMVLFNYMEQQQGGSKVPWDDLKYIFGEIMYG 175
A++ ER++FGP GWN+ Y F DLR S L ++ Q + ++P++ L+Y+ GE YG
Sbjct 3587 ALVQERRKFGPLGWNIPYEFNETDLRISVQQLHMFLNQYE---ELPYEALRYMTGECNYG 3643
Query 176 GHIID 180
G + D
Sbjct 3644 GRVTD 3648
> Hs14730897
Length=1350
Score = 149 bits (375), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 78/184 (42%), Positives = 109/184 (59%), Gaps = 8/184 (4%)
Query 1 QDVVAMQKLDLAHKEGHWVLLQNIHLMPRWTVELEKKLDAFSS--EGPHPNFRCFLSSEL 58
Q +A + + A K G+WV LQN HL W + +E+ + F+ FR FLSS
Sbjct 770 QGPIAEKMVKDAMKSGNWVFLQNCHLAVSWMLAMEELIKTFTDPDSAIKDTFRLFLSSMP 829
Query 59 CDYIPVGILDRSIKLTNEPPQGLQANLKRAFACFPKDDFDEK--DQKVKAIIFGLCFFHA 116
+ PV +L S+K+TNEPP+GL+AN++RAF F+E +K + IIFG+CFFHA
Sbjct 830 SNTFPVTVLQNSVKVTNEPPKGLRANIRRAFTEMTPSFFEENILGKKWRQIIFGICFFHA 889
Query 117 VLLERKRFGPRGWNMNYPFAMGDLRDSAMVLFNYMEQQQGGSKVPWDDLKYIFGEIMYGG 176
++ ERK+FGP GWN+ Y F D + + L Y ++ K+PWD L YI GEI YGG
Sbjct 890 IIQERKKFGPLGWNICYEFNDSDRECALLNLKLYCKE----GKIPWDALIYITGEITYGG 945
Query 177 HIID 180
+ D
Sbjct 946 RVTD 949
> 7301678
Length=4820
Score = 143 bits (361), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 70/181 (38%), Positives = 111/181 (61%), Gaps = 4/181 (2%)
Query 1 QDVVAMQKLDLAHKEGHWVLLQNIHLMPRWTVELEKKLDAFSSEGPHPNFRCFLSSELCD 60
Q+ A++ LD A K+G W++LQN HL+ R+ ELEK LD E PHP+FR +++++
Sbjct 4452 QEKAALRLLDGAIKQGMWLMLQNGHLLIRFVRELEKHLDRI--ENPHPDFRLWITTDPTP 4509
Query 61 YIPVGILDRSIKLTNEPPQGLQANLKRAFACFPKDDFDEKDQ-KVKAIIFGLCFFHAVLL 119
P+GIL +S+K+ EPP GL+ NL+ + ++ + + +++ L FFHAV+
Sbjct 4510 TFPIGILQKSLKVVTEPPNGLKLNLRSTYFKVRQERLESCSHVAFRPLVYVLAFFHAVVQ 4569
Query 120 ERKRFGPRGWNMNYPFAMGDLRDSAMVLFNYMEQQQGGSKVPWDDLKYIFGEIMYGGHII 179
ER+++ GWN+ Y F D +L Y+ + G K+PW+ LKY+ GE+MYGG +I
Sbjct 4570 ERRKYDKLGWNIAYDFNDTDFDVCTEILRTYL-TRCGTGKIPWNSLKYLIGEVMYGGRVI 4628
Query 180 D 180
D
Sbjct 4629 D 4629
> 7298401
Length=4010
Score = 140 bits (352), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 75/185 (40%), Positives = 109/185 (58%), Gaps = 8/185 (4%)
Query 1 QDVVAMQKLDLAHKEGHWVLLQNIHLMPRWTVELEKKLDAFSSEGPHPNFRCFLSSELCD 60
Q +AM+ +D K G+WV+LQN HL + LEK + + HP+FR +L+S D
Sbjct 3440 QGPIAMKMIDEGVKMGNWVVLQNCHLAASFMPLLEKICENLLPDATHPDFRLWLTSYPAD 3499
Query 61 YIPVGILDRSIKLTNEPPQGLQANLKRAFACFPKDDFD-----EKDQKVKAIIFGLCFFH 115
+ PV +L IK+TNEPP+GL++N+ R+ P D + + + K +I+ LCFFH
Sbjct 3500 HFPVVVLQNGIKMTNEPPKGLRSNILRSMISDPISDPEWYESCTQPRIFKQLIYSLCFFH 3559
Query 116 AVLLERKRFGPRGWNMNYPFAMGDLRDSAMVLFNYMEQQQGGSKVPWDDLKYIFGEIMYG 175
AV+ ER+ FGP GWN+ Y F DLR S M L ++ Q + V +D L+Y+ GE YG
Sbjct 3560 AVIQERRYFGPIGWNIPYEFNETDLRISLMQLRMFLNQYE---TVNYDALRYLTGECNYG 3616
Query 176 GHIID 180
G + D
Sbjct 3617 GRVTD 3621
> 7293415
Length=4081
Score = 134 bits (337), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 66/169 (39%), Positives = 101/169 (59%), Gaps = 5/169 (2%)
Query 16 GHWVLLQNIHLMPRWTVELEKKLDAFS--SEGPHPNFRCFLSSELCDYIPVGILDRSIKL 73
GHWV LQN HL + LE + + H +FR +LSS P+ +L S+K+
Sbjct 3511 GHWVFLQNCHLATSFMQTLETIVRNLTLGITKAHVDFRLYLSSMPIQTFPISVLQNSVKI 3570
Query 74 TNEPPQGLQANLKRAFACFPKDDFDEKDQKV--KAIIFGLCFFHAVLLERKRFGPRGWNM 131
TNEPP+G++AN+ A +D F++ Q +AI+FGLC FHAVLLER++FGP GWN+
Sbjct 3571 TNEPPKGIKANVFGALTDLKQDFFEQHIQNGNWRAIVFGLCMFHAVLLERRKFGPLGWNI 3630
Query 132 NYPFAMGDLRDSAMVLFNYMEQQQGGSKVPWDDLKYIFGEIMYGGHIID 180
Y F+ D R+ + ++ ++ ++PW+ + YI G+I +GG + D
Sbjct 3631 TYEFSESD-RECGLKTLDFFIDREVLDEIPWEAILYINGDITWGGRVTD 3678
> Hs22067157
Length=3672
Score = 134 bits (337), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 80/190 (42%), Positives = 107/190 (56%), Gaps = 17/190 (8%)
Query 1 QDVVAMQKLDLAHKEGHWVLLQNIHLMPRWTVELEKKLD-AFSSEGPHPNFRCFLSSELC 59
Q +A + ++ A K+G WV+LQN HL W LEK + E + FR +L+S
Sbjct 3114 QGPIAAKMINNAIKDGTWVVLQNCHLAASWMPTLEKICEEVIVPESTNARFRLWLTSYPS 3173
Query 60 DYIPVGILDRSIKLTNEPPQGLQANLKRAFACFPKDD---FDEKDQKV--KAIIFGLCFF 114
+ PV IL IK+TNEPP+GL+ANL R++ P D F + V + ++FGLCFF
Sbjct 3174 EKFPVSILQNGIKMTNEPPKGLRANLLRSYLNDPISDPVFFQSCAKAVMWQKMLFGLCFF 3233
Query 115 HAVLLERKRFGPRGWNMNYPFAMGDLRDS----AMVLFNYMEQQQGGSKVPWDDLKYIFG 170
HAV+ ER+ FGP GWN+ Y F DLR S M L +Y E VP+D L Y+ G
Sbjct 3234 HAVVQERRNFGPLGWNIPYEFNESDLRISMWQIQMFLNDYKE-------VPFDALTYLTG 3286
Query 171 EIMYGGHIID 180
E YGG + D
Sbjct 3287 ECNYGGRVTD 3296
> Hs15029526
Length=4490
Score = 129 bits (323), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 63/181 (34%), Positives = 107/181 (59%), Gaps = 3/181 (1%)
Query 1 QDVVAMQKLDLAHKEGHWVLLQNIHLMPRWTVELEKKLDAFSSEGPHPNFRCFLSSELCD 60
Q+V A + + ++ ++G WVLLQN HL + EL + L ++E +FR ++++E D
Sbjct 3944 QEVHARKLIQMSMQQGGWVLLQNCHLGLEFMEELLETL--ITTEASDDSFRVWITTEPHD 4001
Query 61 YIPVGILDRSIKLTNEPPQGLQANLKRAFACFPKDDFDEKDQKV-KAIIFGLCFFHAVLL 119
P+ +L S+K TNEPPQG++A LKR FA +D D + + K +++ + F H+ +
Sbjct 4002 RFPITLLQTSLKFTNEPPQGVRAGLKRTFAGINQDLLDISNLPMWKPMLYTVAFLHSTVQ 4061
Query 120 ERKRFGPRGWNMNYPFAMGDLRDSAMVLFNYMEQQQGGSKVPWDDLKYIFGEIMYGGHII 179
ER++FGP GWN+ Y F D S + N++++ V W+ ++Y+ GE+ YG +
Sbjct 4062 ERRKFGPLGWNIPYEFNSADFSASVQFIQNHLDECDIKKGVSWNTVRYMIGEVQYGDRVT 4121
Query 180 D 180
D
Sbjct 4122 D 4122
> 7292540
Length=4680
Score = 125 bits (313), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 66/180 (36%), Positives = 97/180 (53%), Gaps = 9/180 (5%)
Query 5 AMQKLDLAHKEGHWVLLQNIHLMPRWTVELEKKLDAFSSEGPHPNFRCFLSSELCDYIPV 64
A + +++A K G WVLL+N+HL P+W V+LEKK+ + PH FR FL+ E+ +PV
Sbjct 4048 AERAINMACKTGRWVLLKNVHLAPQWLVQLEKKMHSLQ---PHSGFRLFLTMEINPKVPV 4104
Query 65 GILDRSIKLTNEPPQGLQANLKRAFACFPKDDFDEKDQKVKAIIFGLCFFHAVLLERKRF 124
+L EPP G++ANL R F+ P + + + F L +FHA++ ER R+
Sbjct 4105 NLLRAGRIFVFEPPPGIRANLLRTFSTVPAARMMKTPSERARLYFLLAWFHAIVQERLRY 4164
Query 125 GPRGWNMNYPFAMGDLRDSAMVLFNYMEQQQGG------SKVPWDDLKYIFGEIMYGGHI 178
P GW Y F DLR + L +++ G KVPWD L + + +YGG I
Sbjct 4165 VPLGWAKKYEFNESDLRVACDTLDTWIDTTAMGRTNLPPEKVPWDALVTLLSQSIYGGKI 4224
> Hs22049728
Length=2274
Score = 122 bits (307), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 61/180 (33%), Positives = 96/180 (53%), Gaps = 9/180 (5%)
Query 5 AMQKLDLAHKEGHWVLLQNIHLMPRWTVELEKKLDAFSSEGPHPNFRCFLSSELCDYIPV 64
A + ++ A K G WV+L+N+HL P W ++LEKKL + PH FR FL+ E+ +PV
Sbjct 1708 ADKAINTAVKSGRWVMLKNVHLAPGWLMQLEKKLHSLQ---PHACFRLFLTMEINPKVPV 1764
Query 65 GILDRSIKLTNEPPQGLQANLKRAFACFPKDDFDEKDQKVKAIIFGLCFFHAVLLERKRF 124
+L EPP G++AN+ R F+ P + + + F L +FHA++ ER R+
Sbjct 1765 NLLRAGRIFVFEPPPGVKANMLRTFSSIPVSRICKSPNERARLYFLLAWFHAIIQERLRY 1824
Query 125 GPRGWNMNYPFAMGDLRDSAMVLFNYMEQQQGG------SKVPWDDLKYIFGEIMYGGHI 178
P GW+ Y F DLR + + +++ G K+PW LK + + +YGG +
Sbjct 1825 APLGWSKKYEFGESDLRSACDTVDTWLDDTAKGRQNISPDKIPWSALKTLMAQSIYGGRV 1884
> Hs14149700
Length=892
Score = 121 bits (303), Expect = 9e-28, Method: Compositional matrix adjust.
Identities = 62/169 (36%), Positives = 97/169 (57%), Gaps = 5/169 (2%)
Query 14 KEGHWVLLQNIHLMPRWTVELEKKLDAFSSEGPHPNFRCFLSSELCDYIPVGILDRSIKL 73
+ G WV QN HL P W LE+ ++ + + H +FR +L+S + PV IL K+
Sbjct 476 ERGKWVFFQNCHLAPSWMPALERLIEHINPDKVHRDFRLWLTSLPSNKFPVSILQNGSKM 535
Query 74 TNEPPQGLQANLKRAFACFPKDDFDE--KDQKVKAIIFGLCFFHAVLLERKRFGPRGWNM 131
T EPP+G++ANL ++++ +D + K + K+++ LC FH LER++FGP G+N+
Sbjct 536 TIEPPRGVRANLLKSYSSLGEDFLNSCHKVMEFKSLLLSLCLFHGNALERRKFGPLGFNI 595
Query 132 NYPFAMGDLRDSAMVLFNYMEQQQGGSKVPWDDLKYIFGEIMYGGHIID 180
Y F GDLR L ++++ +P+ LKY GEI YGG + D
Sbjct 596 PYEFTDGDLRICISQLKMFLDEYDD---IPYKVLKYTAGEINYGGRVTD 641
> CE23997
Length=4568
Score = 117 bits (293), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 60/176 (34%), Positives = 92/176 (52%), Gaps = 9/176 (5%)
Query 9 LDLAHKEGHWVLLQNIHLMPRWTVELEKKLDAFSSEGPHPNFRCFLSSELCDYIPVGILD 68
L A K G WVLL+N+HL P W +LEK+L + PH FR FL++E+ +P IL
Sbjct 4032 LGTATKSGRWVLLKNVHLAPSWLAQLEKRLHSMK---PHAQFRLFLTAEIHPKLPSSILR 4088
Query 69 RSIKLTNEPPQGLQANLKRAFACFPKDDFDEKDQKVKAIIFGLCFFHAVLLERKRFGPRG 128
S + EP GL+ANL R+ + P + + + +C+ HA++ ER R+ P G
Sbjct 4089 ASRVVVFEPATGLKANLLRSLSSIPPQRLTKAPTERSRLYLLVCWLHALVQERLRYTPLG 4148
Query 129 WNMNYPFAMGDLRDSAMVLFNYMEQQQGG------SKVPWDDLKYIFGEIMYGGHI 178
W+ Y F+ DLR + L ++ G ++PW L+ + + +YGG I
Sbjct 4149 WSTAYEFSDADLRVACDTLDAAVDAVAQGRPNVEPERLPWTTLRTLLSQCIYGGKI 4204
> 7292152
Length=3868
Score = 116 bits (290), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 57/153 (37%), Positives = 88/153 (57%), Gaps = 6/153 (3%)
Query 1 QDVVAMQKLDLAHKEGHWVLLQNIHLMPRWTVELEKKLDAFSSEGPHPNFRCFLSSELCD 60
Q +A + A + G+WV LQN HL W LE + + PNFR +L++
Sbjct 3353 QGPIATALIKNAQEMGYWVCLQNCHLAASWMPYLEYLWENMDTFNTTPNFRIWLTAYPTP 3412
Query 61 YIPVGILDRSIKLTNEPPQGLQANLKRAFACFPKDDFD------EKDQKVKAIIFGLCFF 114
PV IL +K+TNEPP GL+ NL R++ P +D++ ++D+ +++G+CFF
Sbjct 3413 QFPVTILQNGVKMTNEPPTGLKENLMRSYNSEPINDYEFYTGCAKQDRAFTRLLYGICFF 3472
Query 115 HAVLLERKRFGPRGWNMNYPFAMGDLRDSAMVL 147
HAV+ ER+++GP GWN+ Y F DL+ S + L
Sbjct 3473 HAVVQERRKYGPLGWNIAYGFNESDLQISVLQL 3505
> Hs19115954
Length=4624
Score = 116 bits (290), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 60/181 (33%), Positives = 95/181 (52%), Gaps = 3/181 (1%)
Query 1 QDVVAMQKLDLAHKEGHWVLLQNIHLMPRWTVELEKKLDAFSSEGPHPNFRCFLSSELCD 60
Q+V A + L G W LLQN HL + EL + +E H FR ++++E
Sbjct 4079 QEVHARKLLQQTMANGGWALLQNCHLGLDFMDELMDII--IETELVHDAFRLWMTTEAHK 4136
Query 61 YIPVGILDRSIKLTNEPPQGLQANLKRAFACFPKDDFD-EKDQKVKAIIFGLCFFHAVLL 119
P+ +L SIK N+PPQGL+A LKR ++ +D D + K +++ + F H+ +
Sbjct 4137 QFPITLLQMSIKFANDPPQGLRAGLKRTYSGVSQDLLDVSSGSQWKPMLYAVAFLHSTVQ 4196
Query 120 ERKRFGPRGWNMNYPFAMGDLRDSAMVLFNYMEQQQGGSKVPWDDLKYIFGEIMYGGHII 179
ER++FG GWN+ Y F D + + N+++ V W ++Y+ GEI YGG +
Sbjct 4197 ERRKFGALGWNIPYEFNQADFNATVQFIQNHLDDMDVKKGVSWTTIRYMIGEIQYGGRVT 4256
Query 180 D 180
D
Sbjct 4257 D 4257
> Hs13375813
Length=553
Score = 99.0 bits (245), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 59/181 (32%), Positives = 100/181 (55%), Gaps = 11/181 (6%)
Query 1 QDVVAMQKLDLAHKEGHWVLLQNIHLMPRWTVELEKKLDAFSSEGPHPNFRCFLSSEL-C 59
Q +A+Q L + G W+ L+N+HL+ W LEK+L+ P FR +L++E+
Sbjct 5 QADLAIQMLKECARNGDWLCLKNLHLVVSWLPVLEKELNTLQ---PKDTFRLWLTAEVHP 61
Query 60 DYIPVGILDRSIKLTNEPPQGLQANLKRAFACFPKDDFDEKDQKVKA-IIFGLCFFHAVL 118
++ P+ +L S+K+T E P GL+ NL R + + + +KD +A +F L +FHA
Sbjct 62 NFTPI-LLQSSLKITYESPPGLKKNLMRTYESWTPEQISKKDNTHRAHALFSLAWFHAAC 120
Query 119 LERKRFGPRGWNMNYPFAMGDLRDSAMVLFNYMEQQQGGSK-VPWDDLKYIFGEIMYGGH 177
ER+ + P+GW Y F++ DLR +N +++ G+K V W+ + + +YGG
Sbjct 121 QERRNYIPQGWTKFYEFSLSDLRAG----YNIIDRLFDGAKDVQWEFVHGLLENAIYGGR 176
Query 178 I 178
I
Sbjct 177 I 177
> Hs22065713
Length=119
Score = 97.8 bits (242), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 50/112 (44%), Positives = 67/112 (59%), Gaps = 2/112 (1%)
Query 30 WTVELEKKLDAFSSEGPHPNFRCFLSSELCDYIPVGILDRSIKLTNEPPQGLQANLKRAF 89
W L+K ++ E PHP+FR +LSS P+ IL SIK+T EPP+GL+AN+ R +
Sbjct 5 WMPNLDKLVEQLQVEDPHPSFRLWLSSIPHPDFPISILQVSIKMTTEPPKGLKANMTRLY 64
Query 90 ACFPKDDFD--EKDQKVKAIIFGLCFFHAVLLERKRFGPRGWNMNYPFAMGD 139
+ F K K K ++F LCFFH+VLLERK+F GWN+ Y F D
Sbjct 65 QLMSEPQFSRCSKPAKYKKLLFSLCFFHSVLLERKKFLQLGWNIIYGFNDSD 116
> CE05659
Length=4131
Score = 95.5 bits (236), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 59/178 (33%), Positives = 94/178 (52%), Gaps = 11/178 (6%)
Query 1 QDVVAMQKLDLAHKEGHWVLLQNIHLMPRWTVELEKKLDAFSSEGPHPNFRCFLSSELCD 60
Q++ A + + + +G W+ L N+HLM + + K L S PH NFR +L++E
Sbjct 3602 QEIAAYEAIRESASKGEWLCLNNLHLMLQAVPSIFKHL---SLTTPHENFRLWLTTEGDA 3658
Query 61 YIPVGILDRSIKLTNEPPQGLQANLKRAFACFPKDDFDEKDQKVKAIIFGLCFFHAVLLE 120
P +L +S+K+T EPP G++ NL R + D K+ IF L + HA+L E
Sbjct 3659 RFPSMMLQQSLKITFEPPPGVRNNLLRTYTQI---DRSTKNVITCQSIFVLAWLHALLQE 3715
Query 121 RKRFGPRGWNMNYPFAMGDLRDSAMVLFNYMEQQQGGSKVPWDDLKYIFGEIMYGGHI 178
R+ F P+GW Y F D+R V +++E Q +K W+ ++ I ++YGG I
Sbjct 3716 RRTFIPQGWTKFYEFGASDVR----VAKSFVE-QLTANKADWEFVRGILKFVIYGGRI 3768
> Hs22044903
Length=2252
Score = 93.6 bits (231), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 48/126 (38%), Positives = 74/126 (58%), Gaps = 9/126 (7%)
Query 61 YIPVGILDRSIKLTNEPPQGLQANLKRAFACFPKDDFD------EKDQKVKAIIFGLCFF 114
+ PV IL +K+TNEPP GL+ NL +++ P D + K+ + ++FG+CFF
Sbjct 1983 WFPVTILQNGVKMTNEPPTGLRLNLLQSYLTDPVSDPEFFKGCRGKELAWEKLLFGVCFF 2042
Query 115 HAVLLERKRFGPRGWNMNYPFAMGDLRDSAMVLFNYMEQQQGGSKVPWDDLKYIFGEIMY 174
HA++ ERK+FGP GWN+ Y F DLR S L ++ + +P++ + Y+ GE Y
Sbjct 2043 HALVQERKKFGPLGWNIPYGFNESDLRISIRQLQLFINEY---DTIPFEAISYLTGECNY 2099
Query 175 GGHIID 180
GG + D
Sbjct 2100 GGRVTD 2105
> YKR054c
Length=4092
Score = 65.9 bits (159), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 47/178 (26%), Positives = 83/178 (46%), Gaps = 8/178 (4%)
Query 5 AMQKLDLAHKEGHWVLLQNIHLMPRWT-VELEKKLDAFSSEGPHPNFRCFLSSELC-DYI 62
A +++ + EG W+LLQNI + W L K ++ + H F+ F++ L D +
Sbjct 3825 AQEEISKSKIEGGWILLQNIQMSLSWVKTYLHKHVEETKAAEEHEKFKMFMTCHLTGDKL 3884
Query 63 PVGILDRSIKLTNEPPQGLQANLKRAFACFPKDDFDEKDQKVKAI--IFGLCFFHAVLLE 120
P +L R+ + E G+ +K + F K V ++ F L +FHA++
Sbjct 3885 PAPLLQRTDRFVYEDIPGILDTVKDLWGS---QFFTGKISGVWSVYCTFLLSWFHALITA 3941
Query 121 RKRFGPRGWNMNYPFAMGDLRDSAMVLFNYMEQQQGGSKVPWDDLKYIFGEIMYGGHI 178
R R P G++ Y F D + +++ L N + + +PW ++ I+YGG I
Sbjct 3942 RTRLVPHGFSKKYYFNDCDFQFASVYLENVL-ATNSTNNIPWAQVRDHIATIVYGGKI 3998
> Hs22058748
Length=569
Score = 43.9 bits (102), Expect = 2e-04, Method: Composition-based stats.
Identities = 15/21 (71%), Positives = 19/21 (90%), Gaps = 0/21 (0%)
Query 160 VPWDDLKYIFGEIMYGGHIID 180
VPW+D++Y+F EIMYGGHI D
Sbjct 171 VPWEDVRYLFDEIMYGGHITD 191
> SPAC1093.06c
Length=1889
Score = 38.5 bits (88), Expect = 0.007, Method: Composition-based stats.
Identities = 40/167 (23%), Positives = 61/167 (36%), Gaps = 5/167 (2%)
Query 9 LDLAHKEGHWVLLQNIHLMPRWTVELEKKLDAFSSEGPHPNFRCFLSSELCDYIPVGILD 68
L LA E W+ + NIHL W +L K+ S H N R SE+ + +P +L
Sbjct 1643 LKLASTEPLWLFINNIHLSTPWAEKLPSKM----SNHLHKNSRIVCLSEIHNQLPHQLLC 1698
Query 69 RSIKLTNEPPQGLQANLKRAFACFPKDDFDEKDQKVKAIIFGLCFFHAVLLERKRFGPRG 128
S + + NL P + + + F L + HA L E F
Sbjct 1699 ISRSIVFNKQTSFKNNLLNLLELLPTMTHTLPHNRFR-LFFFLSWLHATLAEIYCFTCSS 1757
Query 129 WNMNYPFAMGDLRDSAMVLFNYMEQQQGGSKVPWDDLKYIFGEIMYG 175
W F D +L N + + + W K + ++YG
Sbjct 1758 WKEPCYFDDSDFYFGTKILCNILYRNVHLEEFSWGTFKDLLLNVVYG 1804
> CE17878
Length=2030
Score = 37.7 bits (86), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 18/42 (42%), Positives = 26/42 (61%), Gaps = 2/42 (4%)
Query 12 AHKEGHWVLLQNIHLMPRWTVELEKKLDAFSSEGPHPNFRCF 53
A ++GHW+L+ I+L P L+ + A S+ G HPNFR F
Sbjct 526 AAEKGHWLLVDEINLAPPEC--LDAIVHALSAPGTHPNFRLF 565
> Hs21389555
Length=1021
Score = 36.6 bits (83), Expect = 0.025, Method: Composition-based stats.
Identities = 14/27 (51%), Positives = 17/27 (62%), Gaps = 0/27 (0%)
Query 4 VAMQKLDLAHKEGHWVLLQNIHLMPRW 30
V + L A EGHW++L N HLMP W
Sbjct 405 VVVSTLSQAMYEGHWLVLDNCHLMPHW 431
> Hs13112059
Length=441
Score = 33.5 bits (75), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 23/50 (46%), Gaps = 9/50 (18%)
Query 89 FACFPKDDFDEKDQK--VKAIIFGLCF-------FHAVLLERKRFGPRGW 129
FAC K DF + + ++F +CF F L RK GPRGW
Sbjct 105 FACVVKPDFSTCASRRFLFGVLFAICFSCLAAHVFALNFLARKNHGPRGW 154
> Hs13112057
Length=453
Score = 33.5 bits (75), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 23/50 (46%), Gaps = 9/50 (18%)
Query 89 FACFPKDDFDEKDQK--VKAIIFGLCF-------FHAVLLERKRFGPRGW 129
FAC K DF + + ++F +CF F L RK GPRGW
Sbjct 117 FACVVKPDFSTCASRRFLFGVLFAICFSCLAAHVFALNFLARKNHGPRGW 166
> SPCC737.08
Length=4717
Score = 31.6 bits (70), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 27/52 (51%), Gaps = 10/52 (19%)
Query 12 AHKEGHWVLLQNIHLMPRWTVE-----LEKKLDAFSSE-----GPHPNFRCF 53
A + GHWVLL ++L + +E L+ + +A+ E HPNFR F
Sbjct 1626 AMRNGHWVLLDELNLASQSVLEGLNACLDHRNEAYIPELDKVFKAHPNFRVF 1677
Score = 30.4 bits (67), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 30/60 (50%), Gaps = 16/60 (26%)
Query 12 AHKEGHWVLLQNIHL-----MPRWTVELEKK----LDAFSSEG-------PHPNFRCFLS 55
A +EGHW +L N +L + R LE K ++ ++E PHPNFR FL+
Sbjct 2024 AVEEGHWFVLDNANLCSPAVLDRLNSLLEHKGVLIVNEKTTEDGHPKTIKPHPNFRLFLT 2083
Score = 29.3 bits (64), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 26/55 (47%), Gaps = 13/55 (23%)
Query 12 AHKEGHWVLLQNIHLMPRWTVE-LEKKLDAFSSE------------GPHPNFRCF 53
A + GHWVLL I+L T+E + + L ++ S PH NFR F
Sbjct 635 AVRSGHWVLLDEINLASLETLEPIGQLLSSYESGILLSERGDITPITPHKNFRLF 689
> CE15749
Length=796
Score = 30.0 bits (66), Expect = 2.9, Method: Composition-based stats.
Identities = 25/101 (24%), Positives = 46/101 (45%), Gaps = 18/101 (17%)
Query 11 LAHKEGHWVLLQNIHLMPRWTVELEKK-----------LDAFSS-EGPHPNFRCFLSSEL 58
+AH++ +L+ +I L T+E++KK L +FS + + +F S+++
Sbjct 68 MAHRQRRLLLVTSILLHVASTLEIKKKIFNCFFQTLLSLLSFSKCQARNLSFSITFSNQI 127
Query 59 CDYI------PVGILDRSIKLTNEPPQGLQANLKRAFACFP 93
YI + I L + P+ + N+K F CFP
Sbjct 128 YGYIYNRLKYSLSIDPSKFGLREQAPKTVYVNMKLTFCCFP 168
> Hs22052882
Length=650
Score = 28.9 bits (63), Expect = 5.2, Method: Composition-based stats.
Identities = 12/23 (52%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 56 SELCDYIPVGILDRSIKLTNEPP 78
S LC Y+PVG+L+R + NE P
Sbjct 580 SFLCRYVPVGLLERLPQRINERP 602
> At1g53510
Length=603
Score = 28.5 bits (62), Expect = 8.0, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 24/50 (48%), Gaps = 6/50 (12%)
Query 114 FHAVLLERKRFGPRGWNMNYPFAMGDLRDSAMVLFNYMEQQ--QGGSKVP 161
+H LL+ G G N YP A+G LR F Y+E+ + G +P
Sbjct 347 YHPQLLKDYMSGSEGSNFVYPSAIGHLRQQ----FTYLEENSSRNGPVIP 392
> YMR126c
Length=342
Score = 28.5 bits (62), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 14/49 (28%), Positives = 22/49 (44%), Gaps = 0/49 (0%)
Query 4 VAMQKLDLAHKEGHWVLLQNIHLMPRWTVELEKKLDAFSSEGPHPNFRC 52
+ + DL H+ +LQN+ T+ L+K D +G P RC
Sbjct 85 IPITPADLPHRSSREAVLQNMERSKELTILLKKPKDPVIHDGLEPPRRC 133
Lambda K H
0.325 0.143 0.460
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2806646388
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40