bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2522_orf1
Length=122
Score E
Sequences producing significant alignments: (Bits) Value
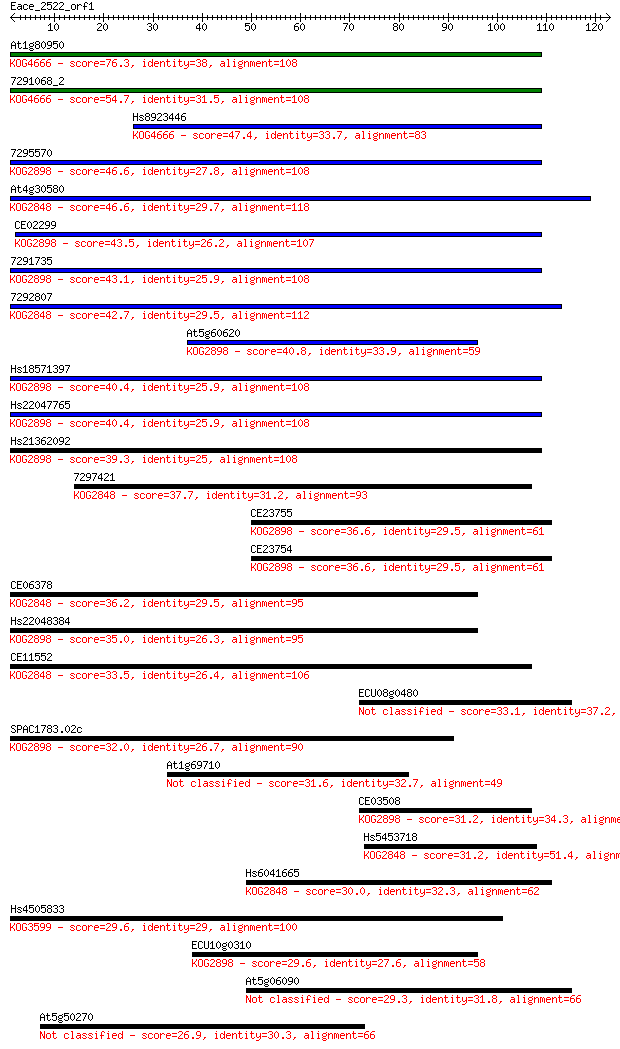
At1g80950 76.3 1e-14
7291068_2 54.7 4e-08
Hs8923446 47.4 6e-06
7295570 46.6 1e-05
At4g30580 46.6 1e-05
CE02299 43.5 1e-04
7291735 43.1 1e-04
7292807 42.7 2e-04
At5g60620 40.8 6e-04
Hs18571397 40.4 9e-04
Hs22047765 40.4 0.001
Hs21362092 39.3 0.002
7297421 37.7 0.006
CE23755 36.6 0.011
CE23754 36.6 0.011
CE06378 36.2 0.015
Hs22048384 35.0 0.031
CE11552 33.5 0.097
ECU08g0480 33.1 0.15
SPAC1783.02c 32.0 0.26
At1g69710 31.6 0.37
CE03508 31.2 0.43
Hs5453718 31.2 0.50
Hs6041665 30.0 1.2
Hs4505833 29.6 1.4
ECU10g0310 29.6 1.4
At5g06090 29.3 1.8
At5g50270 26.9 9.1
> At1g80950
Length=398
Score = 76.3 bits (186), Expect = 1e-14, Method: Composition-based stats.
Identities = 41/109 (37%), Positives = 64/109 (58%), Gaps = 1/109 (0%)
Query 1 HVSCMDIMYFLAQAHAAFVCKASIFHTPLIASPARFLGCISVDRDDAESRRKALEALVS- 59
HVS +DI+Y ++ + +FV K S+ PL+ ++ LGC+ V R+ K + V+
Sbjct 194 HVSYLDILYHMSASFPSFVAKRSVGKLPLVGLISKCLGCVYVQREAKSPDFKGVSGTVNE 253
Query 60 RMKMGSSDTSLNPVVVFPEGTTTNGLGLLRFRRGAFAALQQLQPCVLIY 108
R++ S+ S +++FPEGTTTNG LL F+ GAF A + P +L Y
Sbjct 254 RVREAHSNKSAPTIMLFPEGTTTNGDYLLTFKTGAFLAGTPVLPVILKY 302
> 7291068_2
Length=402
Score = 54.7 bits (130), Expect = 4e-08, Method: Composition-based stats.
Identities = 34/108 (31%), Positives = 51/108 (47%), Gaps = 3/108 (2%)
Query 1 HVSCMDIMYFLAQAHAAFVCKASIFHTPLIASPARFLGCISVDRDDAESRRKALEALVSR 60
H S +D + +A + V K PL+ + I V R+D SR+ + +V R
Sbjct 88 HSSYVDSILVVASGPPSIVAKRETADIPLLGKIINYAQPIYVQREDPNSRQNTIRDIVDR 147
Query 61 MKMGSSDTSLNPVVVFPEGTTTNGLGLLRFRRGAFAALQQLQPCVLIY 108
+ S VV+F EGT TN L++F+ GAF +QP +L Y
Sbjct 148 AR---STDDWPQVVIFAEGTCTNRTALIKFKPGAFYPGVPVQPVLLKY 192
> Hs8923446
Length=427
Score = 47.4 bits (111), Expect = 6e-06, Method: Composition-based stats.
Identities = 28/83 (33%), Positives = 41/83 (49%), Gaps = 3/83 (3%)
Query 26 HTPLIASPARFLGCISVDRDDAESRRKALEALVSRMKMGSSDTSLNPVVVFPEGTTTNGL 85
PLI R + + V R D +SR+ + ++ R G ++VFPEGT TN
Sbjct 54 QVPLIGRLLRAVQPVLVSRVDPDSRKNTINEIIKRTTSGGE---WPQILVFPEGTCTNRS 110
Query 86 GLLRFRRGAFAALQQLQPCVLIY 108
L+ F+ GAF +QP +L Y
Sbjct 111 CLITFKPGAFIPGVPVQPVLLRY 133
> 7295570
Length=407
Score = 46.6 bits (109), Expect = 1e-05, Method: Composition-based stats.
Identities = 30/112 (26%), Positives = 54/112 (48%), Gaps = 11/112 (9%)
Query 1 HVSCMDIMYFLAQAHAAFVCKASIFHTPLIASPARFLGCIS----VDRDDAESRRKALEA 56
H S +D++ + A+ + + HT ++ R L +S DR + R EA
Sbjct 220 HTSPLDVLVLMCDANYSLTGQV---HTGILGVLQRALSRVSHHMWFDRKELADR----EA 272
Query 57 LVSRMKMGSSDTSLNPVVVFPEGTTTNGLGLLRFRRGAFAALQQLQPCVLIY 108
L +++ S PV++FPEGT N +++F++G+FA + P + Y
Sbjct 273 LGLVLRLHCSMKDRPPVLLFPEGTCINNTAVMQFKKGSFAVSDVVHPVAIRY 324
> At4g30580
Length=212
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 35/123 (28%), Positives = 54/123 (43%), Gaps = 11/123 (8%)
Query 1 HVSCMDIMYFLAQAHA-AFVCKASIFHTPLIASPARFLGCISVDRDDAESRRKALEALVS 59
H S +DI L+ + F+ K IF P+I +G + + R D S+ L+ +
Sbjct 58 HQSFLDIYTLLSLGKSFKFISKTGIFVIPIIGWAMSMMGVVPLKRMDPRSQVDCLKRCME 117
Query 60 RMKMGSSDTSLNPVVVFPEGTTTNGLGLLRFRRGAFAALQQ----LQPCVLIYTAGCMHP 115
+K G+S V FPEGT + L F++GAF + + P L+ T M
Sbjct 118 LLKKGAS------VFFFPEGTRSKDGRLGSFKKGAFTVAAKTGVAVVPITLMGTGKIMPT 171
Query 116 AYE 118
E
Sbjct 172 GSE 174
> CE02299
Length=512
Score = 43.5 bits (101), Expect = 1e-04, Method: Composition-based stats.
Identities = 28/107 (26%), Positives = 49/107 (45%), Gaps = 11/107 (10%)
Query 2 VSCMDIMYFLAQAHAAFVCKASIFHTPLIASPARFLGCISVDRDDAESRRKALEALVSRM 61
+SC + + Q A F+ T L S I +R +A R K ++ RM
Sbjct 253 LSCDNCYAMIGQKQAGFL---GFLQTTLSRSEHH----IWFERGEAGDRAKVMD----RM 301
Query 62 KMGSSDTSLNPVVVFPEGTTTNGLGLLRFRRGAFAALQQLQPCVLIY 108
+ +D + P+++FPEGT N ++ F++G+F + P + Y
Sbjct 302 REHVNDENKLPIIIFPEGTCINNTSVMMFKKGSFEIGSTIYPIAVKY 348
> 7291735
Length=537
Score = 43.1 bits (100), Expect = 1e-04, Method: Composition-based stats.
Identities = 28/109 (25%), Positives = 53/109 (48%), Gaps = 5/109 (4%)
Query 1 HVSCMDIMYFLAQAHAAFVC-KASIFHTPLIASPARFLGCISVDRDDAESRRKALEALVS 59
H S +D++ + + + + + F L + AR I +R +A+ R E
Sbjct 334 HTSPIDVLVLMCDSTYSLIGQRHGGFLGVLQRALARASPHIWFERGEAKDRHLVAE---- 389
Query 60 RMKMGSSDTSLNPVVVFPEGTTTNGLGLLRFRRGAFAALQQLQPCVLIY 108
R+K SD + P+++FPEGT N +++F++G+F + P + Y
Sbjct 390 RLKQHVSDPNNPPILIFPEGTCINNTSVMQFKKGSFEVGGVIYPVAIKY 438
> 7292807
Length=343
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 33/118 (27%), Positives = 51/118 (43%), Gaps = 11/118 (9%)
Query 1 HVSCMDIMYFLAQAHA----AFVCKASIFHTPLIASPARFLGCISVDRDDAESRRKALEA 56
H S +D++ H V K +F+ A G I +DR E R+ L
Sbjct 101 HQSSLDVLGMFNIWHVMNKCTVVAKRELFYAWPFGLAAWLAGLIFIDRVRGEKARETLND 160
Query 57 LVSRMKMGSSDTSLNPVVVFPEGTTTNGLGLLRFRRGAF--AALQQLQPCVLIYTAGC 112
+ R+K + VFPEGT N L F++GAF A QQ+ +++++ C
Sbjct 161 VNRRIKKQRIK-----LWVFPEGTRRNTGALHPFKKGAFHMAIDQQIPILPVVFSSYC 213
> At5g60620
Length=376
Score = 40.8 bits (94), Expect = 6e-04, Method: Composition-based stats.
Identities = 20/59 (33%), Positives = 33/59 (55%), Gaps = 4/59 (6%)
Query 37 LGCISVDRDDAESRRKALEALVSRMKMGSSDTSLNPVVVFPEGTTTNGLGLLRFRRGAF 95
+GCI +R +A+ R + L ++ S NP+++FPEGT N + F++GAF
Sbjct 208 VGCIWFNRSEAKDREIVAKKLRDHVQGADS----NPLLIFPEGTCVNNNYTVMFKKGAF 262
> Hs18571397
Length=456
Score = 40.4 bits (93), Expect = 9e-04, Method: Composition-based stats.
Identities = 28/110 (25%), Positives = 51/110 (46%), Gaps = 7/110 (6%)
Query 1 HVSCMDIMYFLAQAHAAFVCKASIFHTPLIASPARFL--GCISVDRDDAESRRKALEALV 58
H S +D++ + + A V + H L+ R + C V + +E + + L A
Sbjct 248 HTSPIDVIILASDGYYAMVGQV---HGGLMGVIQRAMVKACPHVWFERSEVKDRHLVA-- 302
Query 59 SRMKMGSSDTSLNPVVVFPEGTTTNGLGLLRFRRGAFAALQQLQPCVLIY 108
R+ D S P+++FPEGT N ++ F++G+F + P + Y
Sbjct 303 KRLTEHVQDKSKLPILIFPEGTCINNTSVMMFKKGSFEIGATVYPVAIKY 352
> Hs22047765
Length=248
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 28/110 (25%), Positives = 51/110 (46%), Gaps = 7/110 (6%)
Query 1 HVSCMDIMYFLAQAHAAFVCKASIFHTPLIASPARFL--GCISVDRDDAESRRKALEALV 58
H S +D++ + + A V + H L+ R + C V + +E + + L A
Sbjct 40 HTSPIDVIILASDGYYAMVGQV---HGGLMGVIQRAMVKACPHVWFERSEVKDRHLVA-- 94
Query 59 SRMKMGSSDTSLNPVVVFPEGTTTNGLGLLRFRRGAFAALQQLQPCVLIY 108
R+ D S P+++FPEGT N ++ F++G+F + P + Y
Sbjct 95 KRLTEHVQDKSKLPILIFPEGTCINNTSVMMFKKGSFEIGATVYPVAIKY 144
> Hs21362092
Length=434
Score = 39.3 bits (90), Expect = 0.002, Method: Composition-based stats.
Identities = 27/110 (24%), Positives = 50/110 (45%), Gaps = 7/110 (6%)
Query 1 HVSCMDIMYFLAQAHAAFVCKASIFHTPLIASPARFL--GCISVDRDDAESRRKALEALV 58
H S +D++ A V + H L+ R + C V + +E + + L +
Sbjct 229 HTSPIDVLILTTDGCYAMVGQV---HGGLMGIIQRAMVKACPHVWFERSEMKDRHL--VT 283
Query 59 SRMKMGSSDTSLNPVVVFPEGTTTNGLGLLRFRRGAFAALQQLQPCVLIY 108
R+K +D P+++FPEGT N ++ F++G+F + P + Y
Sbjct 284 KRLKEHIADKKKLPILIFPEGTCINNTSVMMFKKGSFEIGGTIHPVAIKY 333
> 7297421
Length=259
Score = 37.7 bits (86), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 29/101 (28%), Positives = 45/101 (44%), Gaps = 17/101 (16%)
Query 14 AHAAFVCKASIFHTPLIASPARFLGCISVDR----DDAESRRKALEALVSRMKMGSSDTS 69
A V K + + P A G + +DR D S +K +A+ R
Sbjct 101 GRATVVSKKEVLYIPFFGIGAWLWGTLYIDRSRKTDSINSLQKEAKAIQERN-------- 152
Query 70 LNPVVVFPEGTTTNGLGLLRFRRGAF-AALQQ---LQPCVL 106
+++FPEGT + LL F++G+F ALQ +QP V+
Sbjct 153 -CKLLLFPEGTRNSKDSLLPFKKGSFHIALQGKSPVQPVVI 192
> CE23755
Length=407
Score = 36.6 bits (83), Expect = 0.011, Method: Composition-based stats.
Identities = 18/63 (28%), Positives = 34/63 (53%), Gaps = 2/63 (3%)
Query 50 RRKALEALVSRMKMGSSDTSLN--PVVVFPEGTTTNGLGLLRFRRGAFAALQQLQPCVLI 107
R +A + LV K+ T+ + P+++FPEGT N ++ F++G+F + P +
Sbjct 171 RSEAKDRLVVAQKLKEHCTNPDKLPILIFPEGTCINNTSVMMFKKGSFEIGTTIYPIAMK 230
Query 108 YTA 110
Y +
Sbjct 231 YDS 233
> CE23754
Length=617
Score = 36.6 bits (83), Expect = 0.011, Method: Composition-based stats.
Identities = 18/63 (28%), Positives = 34/63 (53%), Gaps = 2/63 (3%)
Query 50 RRKALEALVSRMKMGSSDTSLN--PVVVFPEGTTTNGLGLLRFRRGAFAALQQLQPCVLI 107
R +A + LV K+ T+ + P+++FPEGT N ++ F++G+F + P +
Sbjct 381 RSEAKDRLVVAQKLKEHCTNPDKLPILIFPEGTCINNTSVMMFKKGSFEIGTTIYPIAMK 440
Query 108 YTA 110
Y +
Sbjct 441 YDS 443
> CE06378
Length=282
Score = 36.2 bits (82), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 45/97 (46%), Gaps = 7/97 (7%)
Query 1 HVSCMDI--MYFLAQAHAAFVCKASIFHTPLIASPARFLGCISVDRDDAESRRKALEALV 58
H S +DI M + + + K + + P A F I +DR + E +++
Sbjct 98 HQSSLDILSMASIWPKNCVVMMKRILAYVPFFNLGAYFSNTIFIDRYNRERAMASVDYCA 157
Query 59 SRMKMGSSDTSLNPVVVFPEGTTTNGLGLLRFRRGAF 95
S MK + +L + VFPEGT G + F++GAF
Sbjct 158 SEMK----NRNLK-LWVFPEGTRNREGGFIPFKKGAF 189
> Hs22048384
Length=507
Score = 35.0 bits (79), Expect = 0.031, Method: Composition-based stats.
Identities = 25/97 (25%), Positives = 45/97 (46%), Gaps = 7/97 (7%)
Query 1 HVSCMDIMYFLAQAHAAFVCKASIFHTPLIASPARFL--GCISVDRDDAESRRKALEALV 58
H S +D++ + + A V + H LI R + C + + E + + L L
Sbjct 332 HTSPIDVIILASDGYYATVGQ---MHGGLIGLIQRAMVKTCPHIWFERLEVKDRHL--LA 386
Query 59 SRMKMGSSDTSLNPVVVFPEGTTTNGLGLLRFRRGAF 95
R+ D S P+++FPEG N ++ F++G+F
Sbjct 387 KRLTEQVQDISKLPILIFPEGICINNTSVMMFKKGSF 423
> CE11552
Length=262
Score = 33.5 bits (75), Expect = 0.097, Method: Compositional matrix adjust.
Identities = 28/112 (25%), Positives = 47/112 (41%), Gaps = 11/112 (9%)
Query 1 HVSCMDI--MYFLAQAHAAFVCKASIFHTPLIASPARFLGCISVDRDDAESRRKALEALV 58
H S +D+ M F + K+S+ + P A + ++R E K ++ +
Sbjct 94 HQSALDVLGMSFAWPVDCVVMLKSSLKYLPGFNLCAYLCDSVYINRFSKEKALKTVDTTL 153
Query 59 SRMKMGSSDTSLNPVVVFPEGTTTNGLGLLRFRRGAFAALQQ----LQPCVL 106
+ T V ++PEGT LL F++GAF +Q + PCV
Sbjct 154 HEIV-----TKKRKVWIYPEGTRNAEPELLPFKKGAFILAKQAKIPIVPCVF 200
> ECU08g0480
Length=223
Score = 33.1 bits (74), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 24/43 (55%), Gaps = 4/43 (9%)
Query 72 PVVVFPEGTTTNGLGLLRFRRGAFAALQQLQPCVLIYTAGCMH 114
P V+FPEG TN +L+F R ++ C + YT GC++
Sbjct 126 PCVLFPEGCQTNNKAVLQFSRD----VEVDHVCGIRYTGGCIN 164
> SPAC1783.02c
Length=328
Score = 32.0 bits (71), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 24/92 (26%), Positives = 42/92 (45%), Gaps = 8/92 (8%)
Query 1 HVSCMDIMYFLAQAHAAF-VCKASIFHTPLIASPARFLGCISVDRDDAESRRKALEALVS 59
H S +D++ + F VC + + +I++ A F C + K L + +
Sbjct 143 HSSPLDVLVLSCLYNCTFAVCDSKTSNVSIISAQAYFWSCFFSPSKLKITDAKPLAKVAA 202
Query 60 RM-KMGSSDTSLNPVVVFPEGTTTNGLGLLRF 90
+ K+G+ V++FPEG TNG L +F
Sbjct 203 KASKIGTV------VILFPEGVCTNGRALCQF 228
> At1g69710
Length=1028
Score = 31.6 bits (70), Expect = 0.37, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 27/49 (55%), Gaps = 0/49 (0%)
Query 33 PARFLGCISVDRDDAESRRKALEALVSRMKMGSSDTSLNPVVVFPEGTT 81
P R L I D+D+AE L++L++R+K+ T++ P + E T
Sbjct 96 PDRSLDLICKDKDEAEVWVVGLKSLITRVKVSKWKTTIKPEITSAECPT 144
> CE03508
Length=179
Score = 31.2 bits (69), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 72 PVVVFPEGTTTNGLGLLRFRRGAFAALQQLQPCVL 106
PV++FPEG +N +L+FR+ F + P +
Sbjct 9 PVLLFPEGYCSNNSKVLQFRKAIFEENVNIYPVAI 43
> Hs5453718
Length=283
Score = 31.2 bits (69), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 18/36 (50%), Positives = 22/36 (61%), Gaps = 1/36 (2%)
Query 73 VVVFPEGTTTNGLGLLRFRRGAF-AALQQLQPCVLI 107
V VFPEGT + +L F+RGAF A+Q P V I
Sbjct 173 VWVFPEGTRNHNGSMLPFKRGAFHLAVQAQVPIVPI 208
> Hs6041665
Length=278
Score = 30.0 bits (66), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 36/65 (55%), Gaps = 3/65 (4%)
Query 49 SRRKALEALVSRMKMGSSDTSLN-PVVVFPEGTTTNGLGLLRFRRGAF-AALQQLQPCV- 105
+R+++ A+ +G N V ++PEGT + LL F++GAF A+Q P V
Sbjct 142 NRQRSSTAMTVMADLGERMVRENLKVWIYPEGTRNDNGDLLPFKKGAFYLAVQAQVPIVP 201
Query 106 LIYTA 110
++Y++
Sbjct 202 VVYSS 206
> Hs4505833
Length=4302
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 29/100 (29%), Positives = 42/100 (42%), Gaps = 5/100 (5%)
Query 1 HVSCMDIMYFLAQAHAAFVCKASIFHTPLIASPARFLGCISVDRDDAESRRKALEALVSR 60
H + +DI V K ++H SPA FL + V D ++ R A +
Sbjct 3161 HRNSLDIFRIATPHSLGSVWKIRVWHDNKGLSPAWFLQHVIVR--DLQTARSAFFLVNDW 3218
Query 61 MKMGSSDTSLNPVVVFPEGTTTNGLGLLRFRRGAFAALQQ 100
+ S +T N +V E + LLRFRR A LQ+
Sbjct 3219 L---SVETEANGGLVEKEVLAASDAALLRFRRLLVAELQR 3255
> ECU10g0310
Length=451
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 16/58 (27%), Positives = 29/58 (50%), Gaps = 5/58 (8%)
Query 38 GCISVDRDDAESRRKALEALVSRMKMGSSDTSLNPVVVFPEGTTTNGLGLLRFRRGAF 95
G I+ R + R+ +E + + G + P+++FPEGT N + F++G F
Sbjct 168 GSIAFKRSEKIDRQLVVEKVKEHVWSGGA-----PMLIFPEGTCVNNKFSVLFQKGPF 220
> At5g06090
Length=500
Score = 29.3 bits (64), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 34/67 (50%), Gaps = 7/67 (10%)
Query 49 SRRKALEALVSRMKMGSSDTSLNPVVVFPEGTTTNGLGLLRFRRGAFAAL-QQLQPCVLI 107
+R + ++A + + ++ + D +VV+PEGTT LLRF FA L + P +
Sbjct 338 TRIRDVDAEMIKKELSNGD-----LVVYPEGTTCREPFLLRF-SALFAELTDNIVPVAMN 391
Query 108 YTAGCMH 114
Y G H
Sbjct 392 YRVGFFH 398
> At5g50270
Length=423
Score = 26.9 bits (58), Expect = 9.1, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 32/74 (43%), Gaps = 8/74 (10%)
Query 7 IMYFLAQAHAAFVCKASIFHTPLIASPARFLGCIS----VDRDDAESRRKALEALVSR-- 60
+MY L+Q H +C S L + P L + VD + E+ R+ +E ++
Sbjct 343 LMYRLSQHHGHLICSPSPLRDDLSSVPECVLSSLETVEWVDYEGTEAERQLVEFILRNGS 402
Query 61 --MKMGSSDTSLNP 72
K S S+NP
Sbjct 403 CLKKFVISPESVNP 416
Lambda K H
0.327 0.137 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194805952
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40