bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2499_orf1
Length=92
Score E
Sequences producing significant alignments: (Bits) Value
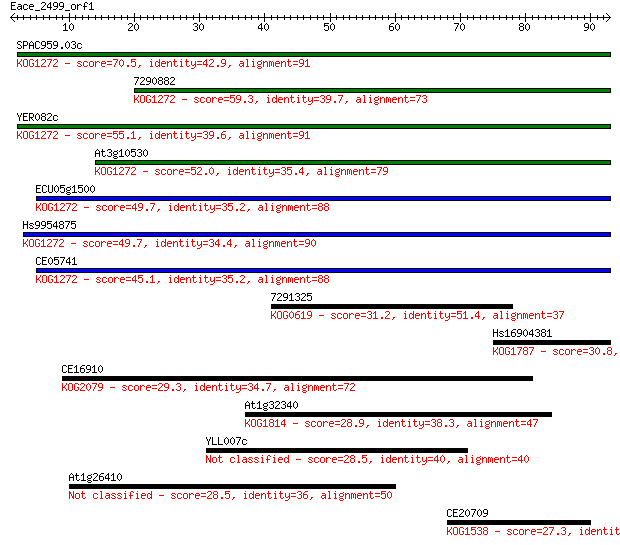
SPAC959.03c 70.5 8e-13
7290882 59.3 2e-09
YER082c 55.1 3e-08
At3g10530 52.0 3e-07
ECU05g1500 49.7 1e-06
Hs9954875 49.7 1e-06
CE05741 45.1 3e-05
7291325 31.2 0.53
Hs16904381 30.8 0.57
CE16910 29.3 2.0
At1g32340 28.9 2.5
YLL007c 28.5 2.8
At1g26410 28.5 3.2
CE20709 27.3 7.0
> SPAC959.03c
Length=520
Score = 70.5 bits (171), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 39/92 (42%), Positives = 56/92 (60%), Gaps = 1/92 (1%)
Query 2 KRSRAIIKQHKTLAAKTKESAAAVSSLFEEQGGYIEAEGLEKVYKFRQEDILKNVSEEIR 61
K+ R+ I++ + + S A L E+ G +EAEGLE+ YKFRQ+ + NV+ E
Sbjct 32 KKLRSNIQKIEERIENVESSLAKTEILHEDNPGLLEAEGLERTYKFRQDQLAPNVALETA 91
Query 62 KKAFRLNLS-YGPYSISCSRDGRYLLLGGDKG 92
K+F L+L +G YS +RDGR +LLGG KG
Sbjct 92 TKSFSLDLDKFGGYSFDYTRDGRMILLGGRKG 123
> 7290882
Length=609
Score = 59.3 bits (142), Expect = 2e-09, Method: Composition-based stats.
Identities = 29/73 (39%), Positives = 44/73 (60%), Gaps = 0/73 (0%)
Query 20 ESAAAVSSLFEEQGGYIEAEGLEKVYKFRQEDILKNVSEEIRKKAFRLNLSYGPYSISCS 79
E AA L +E G ++A+ E +FRQ I +NV + K F LN+ +GPY + +
Sbjct 123 EQAARTELLLQETAGQLQADEGETTAEFRQSQIAENVDIQSSAKHFNLNMEFGPYRMRYT 182
Query 80 RDGRYLLLGGDKG 92
++GR+LLLGG +G
Sbjct 183 KNGRHLLLGGKRG 195
> YER082c
Length=554
Score = 55.1 bits (131), Expect = 3e-08, Method: Composition-based stats.
Identities = 36/93 (38%), Positives = 51/93 (54%), Gaps = 2/93 (2%)
Query 2 KRSRAIIKQHKTLAAKTKESAAAVSSLFEEQGGYIEAEG-LEKVYKFRQEDILKNVSEEI 60
K+ RA +K+ K SAAA L E GY+E E LEK +K +Q +I +V
Sbjct 40 KKLRAGLKKIDEQYKKAVSSAAATDYLLPESNGYLEPENELEKTFKVQQSEIKSSVDVST 99
Query 61 RKKAFRLNL-SYGPYSISCSRDGRYLLLGGDKG 92
KA L+L +GPY I +++G +LL+ G KG
Sbjct 100 ANKALDLSLKEFGPYHIKYAKNGTHLLITGRKG 132
> At3g10530
Length=548
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 45/81 (55%), Gaps = 2/81 (2%)
Query 14 LAAKTKESAAAVSS-LFEEQGGYIEAEGLEKVYKFRQEDILKNVSEEIRKKAFRLNLS-Y 71
L K+ ++AA + L + GY+E EGLEK ++ +Q DI V + + + L +
Sbjct 71 LYGKSAKAAAKIEKWLLPAEAGYLETEGLEKTWRVKQTDIANEVDILSSRNQYDIVLPDF 130
Query 72 GPYSISCSRDGRYLLLGGDKG 92
GPY + + GR++L GG KG
Sbjct 131 GPYKLDFTASGRHMLAGGRKG 151
> ECU05g1500
Length=435
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 31/88 (35%), Positives = 45/88 (51%), Gaps = 0/88 (0%)
Query 5 RAIIKQHKTLAAKTKESAAAVSSLFEEQGGYIEAEGLEKVYKFRQEDILKNVSEEIRKKA 64
R IK K +A + L E GYIEA+ EK Y+ Q+ I ++VS + ++A
Sbjct 7 RERIKNSKKIAEEAHRDVTNYGILRTETCGYIEADSDEKTYEVTQDKIRRHVSIKACEQA 66
Query 65 FRLNLSYGPYSISCSRDGRYLLLGGDKG 92
+ L+ GP SR+G Y+LL KG
Sbjct 67 YNLSFENGPIYAKHSRNGAYMLLRNSKG 94
> Hs9954875
Length=610
Score = 49.7 bits (117), Expect = 1e-06, Method: Composition-based stats.
Identities = 31/91 (34%), Positives = 48/91 (52%), Gaps = 1/91 (1%)
Query 3 RSRAIIKQHKTLAAKTKESAAAVSSLFEEQGGYIEAEGLEKVYKFRQEDILKNVSEEIRK 62
++R+ ++ + +T AA L E+ G++E E E K Q DI++ V
Sbjct 127 KTRSRLEVAEAEEEETSIKAARSELLLAEEPGFLEGEDGEDTAKICQADIVEAVDIASAA 186
Query 63 KAFRLNL-SYGPYSISCSRDGRYLLLGGDKG 92
K F LNL +GPY ++ SR GR+L GG +G
Sbjct 187 KHFDLNLRQFGPYRLNYSRTGRHLAFGGRRG 217
> CE05741
Length=580
Score = 45.1 bits (105), Expect = 3e-05, Method: Composition-based stats.
Identities = 31/96 (32%), Positives = 47/96 (48%), Gaps = 8/96 (8%)
Query 5 RAIIKQHKTLAAKTK-----ESAAAVSSLFEEQGGYI--EAEGLEKVYKFRQEDILKNVS 57
R+ ++Q K K K E+ A E G++ + E Y RQ+DI ++V
Sbjct 95 RSKLQQEKMKVTKDKFQRRIEATAKAEKRLVENTGFVAPDEEDHSLTYSIRQKDIAESVD 154
Query 58 EEIRKKAFRLNL-SYGPYSISCSRDGRYLLLGGDKG 92
K F L L +GPY I + +GR+L++GG KG
Sbjct 155 LAAATKHFELKLPRFGPYHIDYTDNGRHLVIGGRKG 190
> 7291325
Length=267
Score = 31.2 bits (69), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 19/38 (50%), Positives = 24/38 (63%), Gaps = 3/38 (7%)
Query 41 LEKVYKF-RQEDILKNVSEEIRKKAFRLNLSYGPYSIS 77
+EKV +F R +L N + I KKAF LNLS+ Y IS
Sbjct 1 MEKVCQFFRNNYVLANCEDAIYKKAFSLNLSH--YQIS 36
> Hs16904381
Length=2863
Score = 30.8 bits (68), Expect = 0.57, Method: Composition-based stats.
Identities = 12/18 (66%), Positives = 15/18 (83%), Gaps = 0/18 (0%)
Query 75 SISCSRDGRYLLLGGDKG 92
+I SRDG+YLL GGD+G
Sbjct 2784 AIQLSRDGQYLLTGGDRG 2801
> CE16910
Length=1259
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 25/84 (29%), Positives = 40/84 (47%), Gaps = 15/84 (17%)
Query 9 KQHKTLAAKTKESAAAVSSLFEEQGGYI---------EAEGL-EKVYKFRQED--ILKNV 56
K K L K ES + V S FE+ GG + EAE E +Y R+E+ + + +
Sbjct 1110 KYAKQLVDKLLESPSFVESTFEDNGGLLLDIMASCEYEAELYQEMIYLIREENLSLAQKL 1169
Query 57 SEEIRKKAFRLNLSYGPYSISCSR 80
E+ ++A L + P I+C +
Sbjct 1170 EMEVSRRA---PLMHNPNCITCEQ 1190
> At1g32340
Length=688
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 18/47 (38%), Positives = 25/47 (53%), Gaps = 4/47 (8%)
Query 37 EAEGLEKVYKFRQEDILKNVSEEIRKKAFRLNLSYGPYSISCSRDGR 83
E G E +Y++ D L+N S I F + GPY ++CSRD R
Sbjct 302 EQPGQEVLYQW--TDWLQNSS--ISHLGFDNEIVLGPYGVTCSRDKR 344
> YLL007c
Length=665
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 16/46 (34%), Positives = 26/46 (56%), Gaps = 6/46 (13%)
Query 31 EQGGYIEAEGLEKVYKFRQEDILKNVSEEI------RKKAFRLNLS 70
E+G YI +GL+ + F+ EDI ++ E+I RK +NL+
Sbjct 583 EEGRYIWLDGLKLISPFQHEDISEDTKEQIDTLFDLRKNVQMINLN 628
> At1g26410
Length=552
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 18/53 (33%), Positives = 26/53 (49%), Gaps = 3/53 (5%)
Query 10 QHKTL---AAKTKESAAAVSSLFEEQGGYIEAEGLEKVYKFRQEDILKNVSEE 59
Q+ T+ A T+ S A ++ LFE Y+ + E + FR DI N S E
Sbjct 454 QYSTMWFDANATESSLAMMNELFEVAEPYVSSNPREAFFNFRDIDIGSNPSGE 506
> CE20709
Length=1047
Score = 27.3 bits (59), Expect = 7.0, Method: Composition-based stats.
Identities = 11/22 (50%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 68 NLSYGPYSISCSRDGRYLLLGG 89
NL + P+ IS +G YLL+GG
Sbjct 233 NLEFEPHCISYCLNGEYLLIGG 254
Lambda K H
0.313 0.132 0.354
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1171209254
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40